X!Hunter MGF libraries
Mascot 2.6 supports three file formats for spectral libraries: the NIST MSP format, the SpectraST sptxt format and the X!Hunter MGF format. The sptxt format is a variation on MSP and will be covered in a future blog post. The main functional difference between MSP and MGF is in the level of annotation. The X!Hunter MGF format is minimalistic and has only a few match-level items of metadata (sequence, modifications, precursor mass, charge). The peak list is unannotated. In contrast, MSP files usually have a wealth of annotation down to individual peak level.
The Global Proteome Machine (GPM) FTP server contains a number of libraries in X!Hunter MGF format, which have been generated from the GPM search archive. There are no predefined definitions for these, because there are many of them (around 200) and the human and mouse libraries are split into chromosomes. But it’s straightforward to configure an X!Hunter MGF format library in Database Manager. I’ll also show an example of configuring a merged human chromosome library.
Single-file M. tuberculosis H37Rv MGF library
To set up a single-file library, follow the generic steps in the library setup help. I’ve chosen the Mycobacterium tuberculosis strain H37Rv as an example.
- In Database Manager, set up a UniProt proteome database for M. tuberculosis. I used the reference proteome for M. tuberculosis (strain ATCC 25618 / H37Rv).
- Once the database is online, choose Create new in the Library menu.
- Type in a name (e.g. GPM_M._tuberculosis_H37Rv) and choose New custom definition.
- Choose to download files automatically. Paste in the URL to the
file on the GPM FTP server:
ftp://ftp.thegpm.org/projects/xhunter/libs/prokaryotes/mgf/Mycobacterium_tuberculosis_H37Rv_cmp_20.mgf - Click on Create; Next. Edit the filename pattern to contain *.mgf rather than *.msp. Click Start downloading.
- Once downloading is finished, click Edit configuration.
Database Manager suggests suitable parse rules for
accessions,
\([^ ]*\), and descriptions,\(.*\). - Set the MS/MS tolerance to 0.01Da/10ppm.
- Choose the UniProt proteome database as the reference database.
- Configuration is finished, so click on Activate to bring the library online.
The GPM spectra contain the top 20 most intense peaks after averaging a number of observations for the same peptide. The statistics page notes that m/z values have been re-aligned to their exact (calculated) values if a peak is associated with a best-fit b or y fragment. Therefore, it seems safe to set strict MS/MS tolerance.
Concatenated human chromosome MGF library
The GPM human library has one file per chromosome. Use a good FTP client like WinSCP or lftp to download all the files in ftp://ftp.thegpm.org/projects/xhunter/libs/eukaryotes/mgf/human_chromosomes/. Concatenating them into a single file is easiest on the command line and creates a 500MB MGF file. On Windows, use the copy command in a command prompt (cmd.exe):
copy /b *.mgf GPM_human_concatenated_cmp_20.mgf
Equivalent command on Linux is:
cat *.mgf > GPM_human_concatenated_cmp_20.mgf
Each X!Hunter MGF file has three metadata fields at the beginning of the file (SEARCH=, REPTYPE=, LIBSIZE=), which get mixed up when the files are concatenated. However, Mascot ignores these lines, so there is no harm doing so.
Library configuration steps are the same as for the M. tuberculosis steps above. The only differences are: in step 3, choose Upload or copy files manually; in step 4, choose Upload file using web browser; and in step 7, choose Homo sapiens as the taxonomy, or set up a human UniProt proteome database.
It’s possible to roll up all the steps in a simple script and “push” the final file to Database Manager, which may be worth it if the source MGF files are updated often. A previous blog post shows how.
Test search
To verify the libraries are working correctly, I downloaded biological replicate 3 of the whole-cell lysate of M. tuberculosis L7-35 from PRIDE project PXD006117 and peak picked in Mascot Distiller. The choice is mainly of convenience: this is the same data set used in our 2018 ASMS presentation for creating a contaminant library. We know it has some human keratin contamination, so expect to get matches in both libraries.
The M. tuberculosis library contains 150,571 spectra, while the concatenated human library has 1,054,295 spectra. Searching the 65,878 queries of the whole-cell lysate gives 17,744 matches above the default score threshold (300).
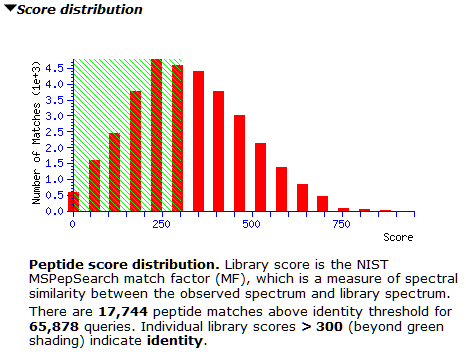
However, the score distribution shows little separation between correct and incorrect matches. It’s a good example for where a decoy library would greatly improve confidence in the results; we’re actively looking into how best to do this. Inspecting matches on both sides of 300 shows many of them have only 3-4 matching peaks, which could be down to the small number of peaks in the library spectra. For now, thresholding at 600 seems prudent to avoid lots of false positives, as the right tail is slightly longer from about 500 onwards.
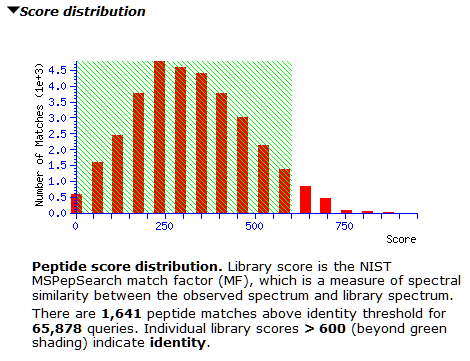
At the new threshold, there are 1,641 strong peptide matches and 256 protein families, each with one family member. Most are tuberculosis proteins as expected, with one hit to human serum albumin. Below are the first few lines from Report Builder:
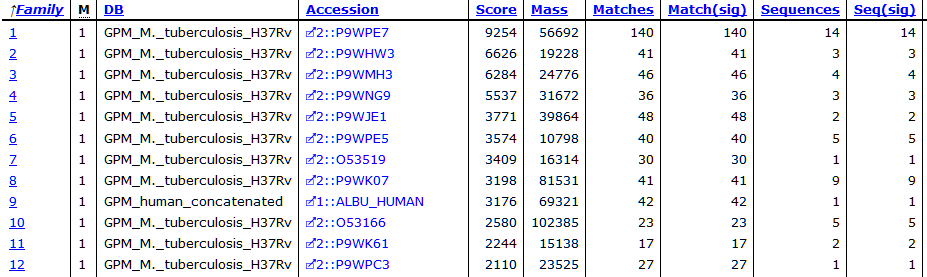
The serum albumin is likely BSA, but because I didn’t include BSA in the search space, it’s not shown here. At score threshold 600, there is no sign of human keratins, although there are matches to them at the lower threshold.
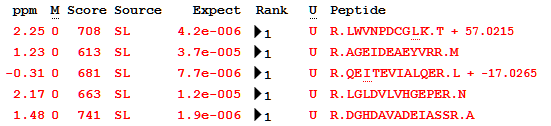
Looking at the list of peptides in one of the tuberculosis proteins (P9WK07, “5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase”), you can see the effect of the minimal library metadata: modifications are simply positive or negative deltas with no other information, as this is all the library provides. This is no obstacle to using X!Hunter MGF files in Mascot, but it is something to keep in mind if you use GPM libraries.
Keywords: database manager, spectral library