Identify proteins by more than ‘gut’ feeling
Last month, we discussed benchmarking protein inference and the role of shared peptide matches. Excluding shared matches may be beneficial to protein identification accuracy if the sequence database contains perfect representations of all proteins in the sample. Many real-life data sets don’t meet this condition. Metaproteomics and environmental samples, such as the various human body sites, peat bog and ocean seawater, are particularly challenging in this respect. These samples cannot be cultivated in the lab, and proteins from different species cannot be separated during sample processing – so, shotgun protein sequencing is done on the mixture proteome.
Shared peptides hold vital information
The protein inference problem is rather more complex than in single-species studies. The software is expected to reconstruct an accurate summary of the mixture proteome of hundreds, if not thousands, of species based on peptide matches to MS/MS spectra. Yet much of the information that links a peptide to a protein in a particular species is lost in sample preparation.
Recall that getting a match to two similar protein sequences could be because the analyte is partly represented by both sequences. This is even more the case in metaproteomics, where nearly all sequence databases are approximate or incomplete (e.g. most microbes lack a reference genome). It is rare for a database to contain the exactly correct sequence variant for any given strain. Additionally, the sample is likely to contain multiple similar bacterial strains as well as closely related species, which means shared peptides are inherent to the data. The Unipept website illustrates this neatly with an interactive database that maps tryptic peptides to microbial species based on UniProt annotations.
The key is to use all the available peptide matches for protein inference, which is what Mascot protein clustering does. Proteins are clustered into families based on shared peptides, and all top-level proteins in the family have at least one unique, significant peptide match. By design, the algorithm isn’t aware of taxonomy, so you will see homologous proteins from different species pulled together. Peptides from analytes without a perfectly representative sequence are put in the same family, and the report displays the maximum number of proteins for which there is distinct evidence, allowing for all the observed shared peptides.
Human gut example
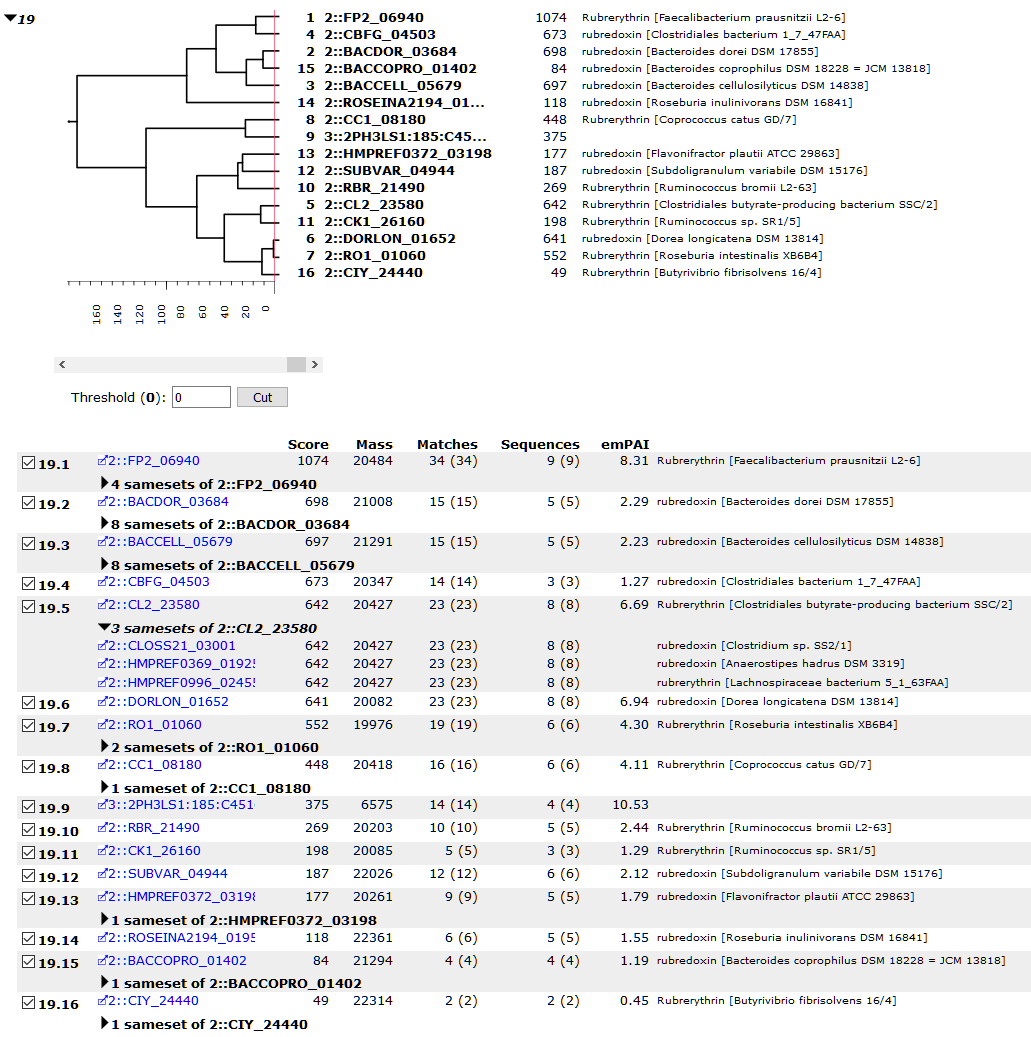
Let’s look at an example from our ASMS 2019 breakfast meeting. This is a search of the data from one subject from a human gut microbiome study. In the screenshot below, database 2 contains translated proteins from the draft genomes of over half of the microbial species (457) identified in the healthy human gut, based on the Human Microbiome Project Reference Genome sequence data. Database 3 is the matched metagenome, which in this study is the translated open reading frames acquired from next-gen sequencing the samples. Results are shown for 1% PSM false discovery rate. (A more detailed description of the database design can be found in the presentation slides.)
The family contains variants of rubredoxin and rubrerythrin from over a dozen species. Sequence similarity between family members ranges from 60% to over 90%, based on a BLAST sequence alignment, while sequence coverage is between 11% (2::CIY_24440) and 55% (2::CL2_23580). The dendrogram reflects the sequence similarity, although it only takes into account observed peptides when computing distance between proteins. In this family, member 19.9 (3::2PH3LS1:185:C451) is the only top-level hit to the matched metagenome. It has one unique peptide match, which confirms that database 2 isn’t complete. The database is clearly missing at least one more rubredoxin or rubrerythrin sequence present in the sample.
Samesets of family member 19.5 are expanded to illustrate that database entries from different species with identical peptide matches are collapsed into the same protein hit. This doesn’t mean the proteins have 100% sequence similarity, just that we can’t tell from the peptide evidence alone which ones are in the sample.
The family has a further 109 subset proteins, most of which have sameset proteins. In total, the subset and “sameset subset” list contains 1294 proteins. These are proteins with no unique peptide matches, so can be explained in terms of the top-level family members. It’s important to keep in mind that absence of evidence isn’t evidence of absence. Any one of the subset proteins could be in the sample, in addition to the top-level proteins, but there is simply no evidence to support this.
There are limits to how much you can infer from shotgun metaproteomics data, as the species-level information is lost in sample processing. Mascot protein clustering seems to work well in this case. Related or similar proteins are pulled together based on shared peptide evidence, then differentiated into family members as much as the data allows. If you’re involved in a metaproteomics study, we’d like to hear from you, so drop us an e-mail.
Keywords: metaproteomics, protein inference