|
To view this email as a web page, click here. |
|
|
|
Welcome
A retrospective on sequence tags shows that they still have their place.
In this month's highlighted publication, peanut allergens are investigated.
The Mascot training course comes with exercises, including de novo sequencing and error tolerant sequence tags.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
30 years of Sequence Tags – and counting
|
|
|
Proteomics has come a long way since spectral interpretation involved using a ruler and paper printouts.
Back in 1994, peptide identification using sequence tags meant printing the MS/MS spectrum on paper and aligning it with ruler lines on a transparent acetate (used with overhead projectors).
The amino acid masses were marked on it to find sequence tags, which were entered manually in a search form and then searched against a sequence database.
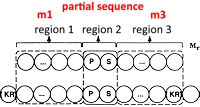
Though modern methods have essentially made the use of sequence tags obsolete, there are still cases where they can help unravel thorny identification problems.
One case is the error tolerant tag (etag), which allows for an unsuspected modification or a small difference in the sequence, e.g. a single nucleotide polymorphism.
When the etag is searched, the peptide molecular weight constraint is relaxed and the fragment ion mass values must fit one of two possibilities.
Either both values are unchanged or both values are shifted by the same amount as the peptide mass.
Error tolerant sequence tag searches are the last step in a search strategy after standard searches, error tolerant searches, and de novo sequencing.
If you have a boutique problem, etags are still a useful technique to have in the toolbox.
There is more history about peptide sequencing at EBML in 1994 in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Proteomic characterization of peanut flour fermented by Rhizopus oryzae
Christopher P. Mattison, Rebecca A. Dupre, Kristen Clermont, John G. Gibbons, Jae-Hyuk Yu
Heliyon 10, e34793 (2024)
Peanuts (Arachis hypogaea) are one of eight foods that commonly cause food allergies, and there are three main immunodominant peanut allergens: Ara h 1, Ara h 2, and Ara h 3. In this study, the authors investigated whether fermentation offers an alternative processing treatment to reduce or eliminate these allergens and make this food safe for peanut-allergic individuals.
The authors fermented peanut flour with Rhizopus oryzae and sampled the mix after 4, 8, 16, 24, and 48 h of fermentation. Proteins were extracted and subject to SDS-PAGE followed by in-gel digestion and LC-MS/MS analysis. The data was searched with a custom Arachis hypogaea database to identify peanut allergens. Mascot Distiller Quantitation Toolbox was used to perform label-free quantitation using the Replicate protocol.
Changes in peptide intensity ratios indicated a decrease in intact allergen proteins relative to unfermented samples. However, the MS of the gel digests still contained significant Ara h 1 and Ara h 3 peptides in samples fermented for 48 h. Additionally, immunoblot analysis showed some reduction in peanut allergens as fermentation time elapsed. The authors concluded that R. oryzae fermented peanut flour under these conditions is not suitable for individuals with peanut allergy.
|

|
|
|
 |
|
|
|
Error tolerant sequence tag exercise
|
|
|
The Mascot Distiller Search Toolbox includes a powerful de novo sequencing algorithm.
The starting point can be any MS/MS scan that has been processed in Distiller to create a peak list.
Short sequence tags generated by the algorithm can be submitted to Mascot Server for a sequence query search to pull out the right peptide(s) from the database.
The training course module Sequence Queries has more details and an example.
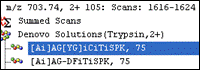
Error tolerant sequence tags are of course supported.
Exercise SQ3 uses an etag to identify an unknown modification in a peptide.
Hints and answers are provided, but it's worth doing the exercise yourself to see how useful an etag can be.
Please contact us for a free 30-day licence for Mascot Distiller.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|