|
To view this email as a web page, click here. |
|
|
|
Welcome
Mascot Server 3.0 is in beta testing. One of the new capabilities is utilising predicted retention time and fragmentation spectra in boosting peptide identifications.
In this month's highlighted publication, the authors have developed a method for the untargeted identification of viruses.
Update your licence now and get Mascot Server 3.0 on release.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Refining results with machine learning
|
|
|
Mascot Server 3.0, currently in beta testing, incorporates the latest computational advances in retention time and fragment intensity predictions.
The predicted features are used in separating correct and incorrect matches, which boosts peptide and protein identifications.
The predictions are provided by MS2Rescore, which is integrated directly with Mascot Server.
MS2Rescore is a "Modular and user-friendly platform for AI-assisted rescoring of peptide identifications" developed at the University of Ghent.
It includes two prediction systems.
DeepLC is a "retention time predictor for (modified) peptides that employs Deep Learning", while MS2PIP provides "Fast and accurate peptide fragmentation spectrum prediction for multiple fragmentation methods, instruments and labeling techniques".
Unlike many other ML models, they are able to make accurate predictions for variable modifications not seen in the training step.
We selected a small spiked yeast data set as an example and searched with Mascot.
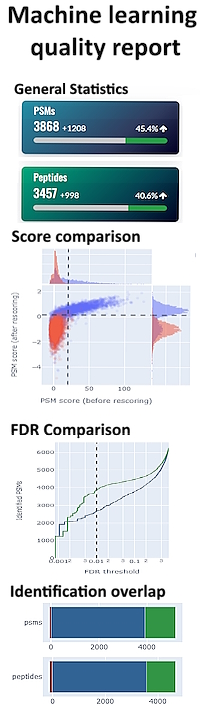
Mascot Server 3.0 with MS2Rescore identified 17% more unique sequences and 13% more protein hits than fully optimised Mascot Server 2.8.
To help give you confidence in your results, Mascot includes a machine learning quality report, which helps answer questions like: Did I choose the right model? Are the predictions any good?
Read more about the upcoming new machine learning capabilities of Mascot Server 3.0.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Universal Identification of Pathogenic Viruses by liquid chromatography coupled to tandem mass spectrometry Proteotyping
Clément Lozano, Olivier Pible, Marine Eschlimann, Mathieu Giraud, Stéphanie Debroas, Jean-Charles Gaillard, Laurent Bellanger, Laurent Taysse, Jean Armengaud
Molecular & Cellular Proteomics, published online July 29, 2024
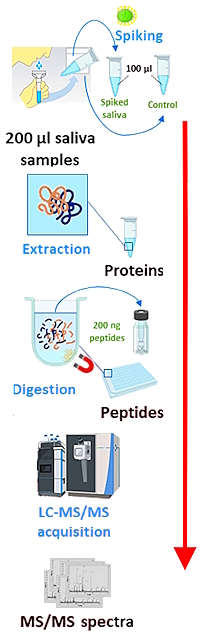
The authors have developed a method for the untargeted identification of viruses in samples. The method involves protein extraction from saliva samples using magnetic beads, followed by digestion and LC-MS/MS.
Taxonomical identification was performed using the Mascot search engine and a cascade search. In the first round, the 10,000 best MS/MS spectra were selected and queried against a subset of the NCBInr database. This subset comprises 50,995 organisms with one representative per species (494 Archaea, 2231 Eukaryota, 12,047 Bacteria, and 36,223 Viruses). During the second round, the database was reduced to the genera identified during the first round and all their descendants, and all MS/MS spectra were searched against the reduced database. The last round consisted in the repeat search of all MS/MS spectra against a database reduced to the species identified during the second search.
As a proof of concept, the authors assessed the method on publicly available shotgun proteomics datasets obtained from virus preparations of infected individuals. Successful virus identification was achieved with 53 public datasets, spanning 23 distinct viral species. The saliva samples collected from four healthy donors were spiked with the vaccinia virus and analyzed with the method, showing the presence of the virus in all and with no false positives. The method was also able to discriminate between three closely related alphaviruses, namely Pixuna virus, Rio Negro virus and the Sindbis virus. Overall, the method displayed a sensitivity of 87.9% and a specificity of 79.6%.
|
|
|
|
 |
|
|
|
Update now and get Mascot Server 3.0 on release
|
|
|
Mascot Server 3.0 is in beta testing, and we aim to release it by the end of September.
Customers with an active Premium Support contract will receive the new version on release for free.
See what's new in Mascot Server 3.0.
If you have an older Mascot Server licence,
send us your product key
by e-mail and we'll quote you for a version update, which comes with 1 year of Premium Support.
Updating before 3.0 has been released is more cost effective than waiting until after the release.
The offer expires on 30 September but quotes are valid for 60 days – act now to secure your price.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|