|
To view this email as a web page, click here. |
|
|
|
Welcome
Mascot Server 3.0 is released! With machine learning, improved error tolerant searching as well as new Mascot Daemon 3.0, there is much to be excited about.
In this month's highlighted publication, the authors use thermal profiling and affinity selection to determine natural product binders and protein targets.
Are you attending the HUPO World Congress in Dresden, Germany on October 20-24? Stop by our booth for a live demo of Mascot Server 3.0.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Mascot Server 3.0
|
|
|
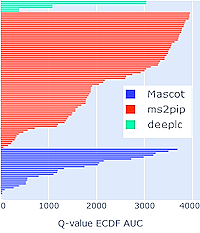
Mascot Server 3.0 brings you powerful new capabilities for AI-assisted rescoring of peptide identifications, resulting in improved coverage and sensitivity.
The MS2Rescore integration includes DeepLC for retention time prediction and MS2PIP for predicting MS2 fragmentation spectra. Both tools have been proven to increase the sensitivity of database search results in all types of experiment, particularly – but not limited to – endogenous peptides, proteogenomics and metaproteomics.
Error tolerant searching has been upgraded. This is a two-pass search that identifies unsuspected chemical and post-translational modifications as well as enzyme non-specificity.
You can now select specific modification classes for the second pass to constrain the search space. For example, search only post-translational modifications, or only N-linked glycosylation.
Mascot Server 3.0 introduces a new file format, Mascot Search Results (MSR). This is a self-contained SQLite file optimised for read performance. Browsing and exporting search results is quicker and more responsive.
Every aspect of the system has been re-engineered under the hood, but Mascot Server continues to have strong backwards compatibility. All user documentation has been reviewed, and we have added dozens of new help pages and tutorials.
Read about more of the capabilities of Mascot Server 3.0, and get a quote today.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Integrated Thermal Proteome Profiling and Affinity Ultrafiltration Mass Spectrometry (iTPAUMS): A Novel Paradigm for Elucidating the Mechanism of Action of Natural Products
Hengyuan Yu, Yang Chen, Yichen Wang, Weiliang Fu, Rui Xu, Jie Liu, Yong Chen, Xuesong Liu, Yongjiang Wu, and Tengfei Xu
Analytical Chemistry, Articles ASAP, published online: September 10, 2024
With over 60% of the small-molecule therapeutics approved by the FDA in the last ~40 years having their roots in natural products, the authors sought to develop a more efficient way to screen for new targets and modes of action.
They first employed thermal proteome profiling (TPP) leveraging ligand-induced thermal stabilization to detect ligand−protein interactions. This is followed by the expression and purification of each direct target protein for subsequent affinity ultrafiltration mass spectrometry (AUMS) screening to establish specific one-to-one protein ligand interactions.
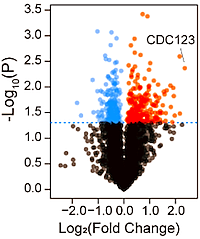
To test their process, the authors incubated a cell lysate with hemp oil extract (HOE) and DMSO (control) at various temperatures. The protein was subsequently trypsinated, labeled with TMT agents, mixed, and analyzed by LC-MS/MS. By comparing the changes in soluble protein content before and after HOE treatment, they found an increase in the overall thermal stability of proteins. After HOE treatment, 16,351 peptides, 3,250 protein groups, and 2,725 quantified protein groups were identified.
From the fold change and p-value, they selected the protein CDC123 as a good target, prepared the protein, and incubated it with the HOE. After ultrafiltration separated the combined ligand−protein complexes from the unbound compounds, the ligands released from the complexes were identified by LC-MS/MS. The ligand exhibiting the most substantial binding affinity was identified as Cannabidiol (CBD).
|
|
|
|
 |
|
|
|
Mascot at HUPO 2024
|
|
|
HUPO World Congress 2024 is held in Dresden, Germany on October 20-24.
Come and meet Matrix Science at booth #21 in the industry exhibition area.
See a live demo of new features in Mascot Server 3.0, ask for advice, find out how to upgrade your licence.
The best time to pop by is during the poster sessions, 11:45-13:00 and 15:35-16:30, as well as during the welcome reception on the opening night.
Please e-mail us if you'd like to meet at a specific time during the conference.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|