|
To view this email as a web page, click here. |
|
|
|
Welcome
We reanalyse a data set that studies endogenous proteolytic processing, and compare the results to FragPipe.
This month's highlighted publication provides comprehensive characterization of noncanonical peptides from human gastric cancer-related samples.
Mascot Server 3.1 is now available. This patch release enhances integration with Thermo Proteome Discoverer™ and fixes a few important bugs introduced in Mascot Server 3.0.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
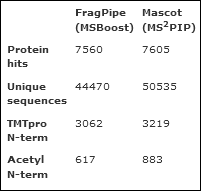
Uncovering endogenous proteolytic processing (Mascot vs FragPipe)
|
|
|
Studying endogenous proteolytic processing, or N-terminomics, involves database searching of semi-specific peptides.
Mascot Server 3.0 includes MS2PIP machine learning models for fragment intensity prediction, which can give a big boost to semi-specific identifications.
We reanalysed data from a recent publication, deposited on MassIVE, where the authors process TMTpro-labelled samples from a terminomic murine polycystic kidney disease (PDK) model.
The TMTpro labelling was performed at the protein level, so N-terminal labels must be treated as variable mods.
We also used a custom enzyme to model the cleavage rules, as trypsin cannot cleave at the TMTpro-labelled lysines.
Mascot ships with an MS2PIP model for TMT-labelling, which is a good fit to these spectra.
Just over 7,600 proteins were identified at 1% sequence FDR, which includes 3,219 matches to TMTpro N-term and 883 to Acetyl N-term.
Recent versions of FragPipe follow the same rescoring idea as MS2PIP, with MSBoost providing the predictions.
It turns out, Mascot + MS2PIP find at least as many protein hits as FragPipe + MSBoost, but also about 13% more unique peptide sequences at 1% FDR.
The searches were done against the mouse proteome (63k entries), which is tiny relative to what Mascot is designed to do.
The semi-tryptic Mascot search takes just 5 minutes on a recent 16-core PC, and machine learning takes a few more minutes at the end.
The full details can be found in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Comprehensive discovery and functional characterization of the noncanonical proteome
Chengyu Shi, Fangzhou Liu, Xinwan Su, Zuozhen Yang, Ying Wang, Shanshan Xie, Shaofang Xie, Qiang Sun, Yu Chen, Lingjie Sang, Manman Tan, Linyu Zhu, Kai Lei, Junhong Li, Jiecheng Yang, Zerui Gao, Meng Yu, Xinyi Wang, Junfeng Wang, Jing Chen, Wei Zhuo, Zhaoyuan Fang, Jian Liu, Qingfeng Yan, Dante Neculai, Qiming Sun, Jianzhong Shao, Weiqiang Lin, Wei Liu, Jian Chen, Liangjing Wang, Yang Liu, Xu Li, Tianhua Zhou & Aifu Lin
Cell Research (2025) doi:10.1038/s41422-024-01059-3
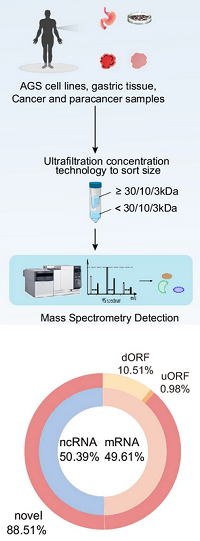
Novel peptidomics could facilitate the understanding of diseases like gastric cancer.
Starting from human transcript data from Ensembl, the authors created a reference database of over 11 million potential sORFs.
They prepared several samples of cancer/paraneoplastic tissue, normal gastric tissue and AGS gastric cancer cell lines.
The samples were milled and lysed, then analysed with ultrafiltration tandem MS.
Database searches with Mascot yielded 8,495 matches to previously unannotated peptides among the 11 million sORFs.
Of these, 4,866 were supported by at least two PSMs and 2,290 were detected across different samples.
Bioinformatic characterisation showed the novel peptide genes were mostly localised to chromosomes 17, 19 and 22.
The authors used CRISPR screening to identify 1,161 peptide candidates that influence AGS cell proliferation, the majority of which appear to function as pro-proliferative regulators.
They also selected 100 peptides with significant phenotypic effects identified from the CRISPR screening for function prediction using AlphaFold2 and peptide-protein interaction networks.
Finally, the authors have established the Human Novel Peptides Atlas Database to encourage further characterisation and validation of the novel peptidome.
|
|
|
|
 |
|
|
|
Mascot Server 3.1
|
|
|
We are pleased to announce the release of Mascot Server 3.1.
Product keys and download links have been sent to customers under support.
Please contact us if you haven't received the e-mail or if you'd like to update.
The client API has been enhanced in two ways.
You can now define machine learning parameters as part of the instrument definition; for example, select a specific MS2PIP model for your instrument.
When active, Mascot runs machine learning after the database search and sends the refined results to the client software.
This is automatically available to any software using the 'result_file_mime' API call, including all versions of Thermo Proteome Discoverer™.
No software alterations are needed.
This patch release also resolves a few bugs and issues reported in Mascot Server 3.0, so we advise all users to update.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|