|
To view this email as a web page, click here. |
|
|
|
Welcome
Join us at the upcoming Matrix Science User Meeting on 2 June at ASMS 2025, Baltimore, Maryland.
Are you presenting a poster or talk at ASMS that uses Mascot? Send us the poster/talk number, presenter name and abstract to newsletter@matrixscience.com and we will feature it in a future newsletter.
This month's highlighted publication uses zero-length crosslinking to probe the interaction landscape between GPIHBP1 and lipoprotein lipase.
Use the CID and HCD models shipped with Mascot to boost your identifications.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Matrix Science ASMS User Meeting
|
|
|
Please join us at the upcoming ASMS User Meeting held on 2 June in Baltimore, Maryland.
There is no charge for attending the meeting, but advance registration is required. Numbers will be limited, so early registration is advisable. Breakfast will be provided.
Monday 2 June, 7:00 am – 8:00 am
Room 338, Baltimore Convention Center, Baltimore, MD
Using MS2Rescore to boost identification rates in complex samples,
Tim Van Den Bossche, Computational Omics and Systems Biology Group (CompOmics), Ghent University/VIB-UGent Center for Medical Biotechnology Mascot DIA: Universal spectrum-centric approach,
Ville Koskinen, Matrix Science Mascot DIA: Thermo LFQ example,
Patrick Emery, Matrix Science
Register now
|

|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
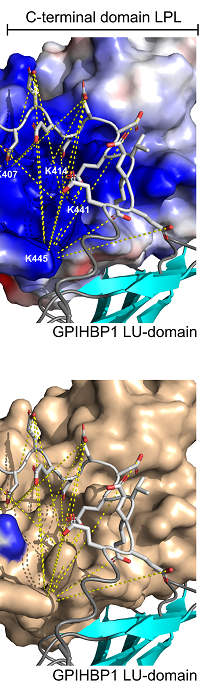
Competitive displacement of lipoprotein lipase from heparan sulfate is orchestrated by a disordered acidic cluster in GPIHBP1
Anamika Biswas, Samina Arshid, Kristian Kølby Kristensen, Thomas J.D. Jørgensen, Michael Ploug
Journal of Lipid Research, 66(2):100745, 2025
Elevated plasma levels of triglyceride-rich lipoproteins (TRLs) and their cholesterol-enriched remnant particles are implicated in the risk of acute pancreatitis and atherosclerotic cardiovascular disease.
Rapid conversion of TRLs depends on a highly regulated flux of active lipoprotein lipase (LPL).
The shuttling of interstitial LPL is done by GPIHBP1, the obligate endothelial trafficking receptor.
The authors clarify the molecular mechanisms by which GPIHBP1 dislodges LPL from its confinement on heparan-sulfate proteoglycans.
In addition to differential scanning fluorimetry, lipase activity assays and hydrogen-deuterium exchange mass spectrometry, they used zero-length crosslinking to provide an experimental mapping of the interaction landscape for the acidic tail of GPIHBP1 on human LPL (hLPL).
Recombinant and secreted versions of human GPIHBP1 were crosslinked with hLPL using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC).
After separation with SDS-PAGE, the excised crosslinked products were digested, 18O labelled and analysed with LC-MS/MS.
The raw data were processed with Mascot Distiller.
Crosslinked peptides were then identified with Mascot Server, allowing for several variable modifications:
methionine oxidation, pyroglutamate formation at glutamic acid, sulfation at tyrosine and (for labelled samples), 18O label at C-terminus.
They identified 16–22 unique crosslinks between hLPL and the acidic tail of GPIHBP1 with Mascot score ≥80, which were validated by the increased incorporation of C-terminal 18O label.
The acidic residues in GPIHBP1 mapped close to the interface between the C- and N-terminal domains in LPL, which revealed widespread polyelectrolyte interactions spanning both LPL domains.
|
|
|
|
 |
|
|
|
Best MS2PIP model for Thermo Orbitrap
|
|
|
For qualitative work and label-free quantitation with Thermo Orbitrap instruments, Mascot Server ships with three MS2PIP models: CID, HCD2019/HCD2021 and Immuno-HCD.
When enabled, MS2PIP predicts the MS/MS spectra of identified peptides, and the correlation between observed and predicted can be used for boosting correct identifications and penalising incorrect ones.
This almost always improves the count of identified peptides without increasing the false discovery rate.
The CID model, as the name implies, is best with linear ion traps and in-source CID, while HCD2021 model is best with HCD.
For immunopeptides, Mascot also ships with the MS2PIP model Immuno-HCD. This model is intended for HLA-I and HLA-II peptides, although it has good performance with tryptic digests too.
More details are in our blog, including a comparison between Mascot Server 2.7, 2.8 and 3.0.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|