|
To view this email as a web page, click here. |
 |
|
Welcome
Putting your Mascot Server on Amazon's cloud is very easy, but is it a good idea?
We look at the important pros and cons.
This month's highlighted publication shows a method to improve reproducibility of peptide analysis
with the use of chip-based multi-phase chromatography coupled to MS/MS.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is using Mascot Daemon to automate the export of search results.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Hosting Mascot Server in "The Cloud"
There's a lot of hype surrounding cloud computing. We've taken a hard-nosed look at the pros and cons of putting your Mascot Server
on the cloud using Amazon Web Services.
Executive summary:
- Performance - Processor speed is excellent. I/O is also very fast: less than 7 minutes to download a 15GB Fasta file from NCBI.
- Security - Can be as good or better than hosting a server in-house. Using appropriate firewall settings and
regularly backing up your data are the critical factors, whether in-house or on the cloud.
- Cost - May be higher or lower than using in-house hardware. A lot depends on whether you are charged overheads for power, rack space, etc.
Perhaps the greatest advantage of using the cloud is convenience. We've written scripts for
AWS CloudFormation that automate the building of a virtual server based on disk images for Mascot Server 2.5.1 pre-installed and
configured with a selection of popular databases. You really can be up and running in just a few minutes.
For more information, see
this presentation from our recent ASMS user meeting. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
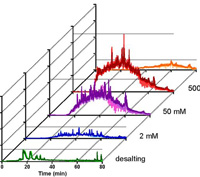
Online Peptide Fractionation Using a Multiphasic Microfluidic Liquid Chromatography Chip Improves Reproducibility and Detection Limits for Quantitation in Discovery and Targeted Proteomics.
Christoph Krisp, Hao Yang, Remco van Soest, and Mark P Molloy
Molecular & Cellular Proteomics (2015) 14: 1708-1719.
This paper presents separation methods for improved reproducibility, which is often required for the workflows used in high-throughput comparative proteomics. The LC chip format enables fast workflow switching for easier transition from discovery to targeted studies, and provides a way to help standardize cross-laboratory proteomic analyses.
In the data-dependent MS/MS analysis of a cancer cell lysate 95% of all identified peptides were uniquely identified in a single fraction and less than 0.9% in more than two fractions. Using targeted detection with scheduled MRM high resolution MS, 503 peptides representing 173 plasma proteins were analyzed. In this multiphase fractionation approach all 503 peptides were quantifiable with a median CV of 3.4 +/- 3%, with only 10 peptides showing CVs > 15%. Three thyroid cell lines were analyzed with multiphase LC chip fractionation via data-independent MS/MS resulting in a total of 3875 proteins identified. You can access these data sets at ProteomeXchange.
|
 |
 |
 |
 |
|
Mascot tip of the month
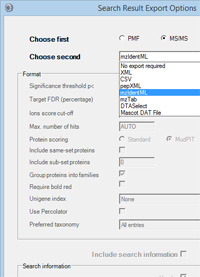
Mascot Daemon 2.5 provides an easy way to automatically export a batch of search results.
On the task editor tab, there's a new button for "Auto-export". Click this button and you can select the export file format that you want. Different formats have different options, so make sure that you choose the type of search that you are doing first, followed by the export format. The exported files are saved in the task folder - normally in a subdirectory under: C:\ProgramData\Matrix Science\Mascot Daemon\MGF. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|