|
To view this email as a web page, click here. |
 |
|
Welcome
Finding a non-specific modification is just one of the 10 Mascot tips & tricks that we've collected together into a new document called "How do I do that?"
This month's highlighted publication shows a new approach to selectively analyze glycoproteins from human plasma.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
The videos in the webcast training course have recently been updated.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
How Do I Do That?
We collected together 10 useful Mascot tips & tricks for a presentation at our recent ASMS User Meeting. Here are just three of the topics:
- Find a non-specific modification - You might be tempted to create a modification such as FuzzyMod
(ACDEFGHIKLMNPQRSTVWY). If you try to use such a modification in a search,
Mascot will almost certainly run out of memory or address space and crash because of
the combinatorial explosion. What you need to do is ...
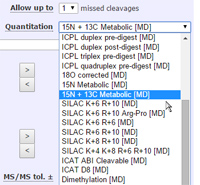
- Metabolically labelled samples - To search data from metabolically labelled samples, e.g. 15N or 13C data, you select a quantitation method, even though you might not be interested in performing quantitation.
- Free up disk space on your Mascot Server - No matter how big your disk, it will eventually fill up. You can download a script that will recover space by periodically deleting old cache files and compressing result files.
To read more about these and the 7 other topics, see
How Do I Do That? |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Simultaneous Enrichment of Plasma Soluble and Extracellular Vesicular Glycoproteins Using Prolonged Ultracentrifugation-Electrostatic Repulsion-hydrophilic Interaction Chromatography (PUC-ERLIC) Approach
Esther Sok Hwee Cheow, Kae Hwan Sim, Dominique de Kleijn, Chuen Neng Lee, Vitaly Sorokin and Siu Kwan Sze
Molecular & Cellular Proteomics (2015) 14: 1657-1671.
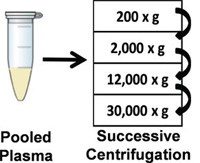
The authors investigated circulating glycoproteins and extracellular vesicles secreted from damaged or infected host cells and tissues, potential sources of biomarkers of human pathologies. The study showed that glycoprotein enrichment by PUC-ERLIC facilitates the simultaneous recovery of both secretory and extracellular vesicle-derived glycoproteins from non-depleted human plasma.
In a single LC-MS/MS run, the authors identified 127 glycoproteins in human plasma samples, including 599 unique glycopeptides and 361 unique glycosylation sites. The PUC-ERLIC method enabled the detection of 48 low-abundant glycoproteins (pg/ml-ng/ml) in human plasma without the need for prior depletion of high-abundance molecules. Using plasma obtained from heart disease patients, they identified, LYVE1, a cardiac-specific remodeling glycoprotein that has been reported to be involved in myocardium healing.
|
 |
 |
 |
 |
|
Updated training course
Whether you are new to database searching or an expert looking for specific information, the webcast training course modules contain a great deal of useful information that complements the material in the help pages. All modules have recently been updated to reflect new functionality in Mascot 2.5.
Follow this link to go directly to the online videos.
When you've finished, there is a multiple choice examination. Achieve an accuracy of 75% or better, and you get to print out a personalised certificate of completion. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|