|
To view this email as a web page, click here. |
 |
|
Welcome
Do you have instruments from more than one vendor? Mascot Distiller provides a one-stop solution for high quality peak picking, de novo sequencing, quantitation, and much more.
This month's highlighted publication features a method to rapidly type E. coli strains at the subspecies level by MALDI-TOF MS.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to customize modification settings, such as the contents of the short list of common modifications.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Streamline your data processing with Mascot Distiller
There are many useful functions packed into Mascot Distiller - raw data browser, high quality peak picking, de novo sequencing, MS1 quantitation, search result viewer - we could go on. Perhaps the most powerful aspect is that it works with raw data from all the major vendors: Agilent, Bruker, Sciex, Shimadzu, Thermo and Waters. If you have instruments in your laboratory from more than one vendor, being able to use one piece of software across all the data formats is a major time-saver.
You can take advantage of the free 30-day trial to discover how Distiller can streamline your data processing.
There is a brand new tutorial to help you get up to speed with peak picking. Or if you prefer, you can just send us a typical raw file and we'll return a suitable set of processing settings. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
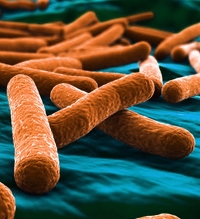
Rapid, Sensitive, and Specific Escherichia coli H Antigen Typing by Matrix-Assisted Laser Desorption Ionization–Time of Flight-Based Peptide Mass Fingerprinting.
Huixia Chui, Michael Chan, Drexler Hernandez, Patrick Chong, Stuart McCorrister, Alyssia Robinson, Matthew Walker, Lorea A. M. Peterson, Sam Ratnam, David J. M. Haldane, Sadjia Bekal, John Wylie, Linda Chui, Garrett Westmacott, Bianli Xu, Mike Drebot, Celine Nadon, J. David Knox, Gehua Wang and Keding Cheng
J. Clin. Microbiol. (2015), vol. 53, 2480-2485
This paper describes a rapid method to identify the Escherichia coli flagellar antigen (H antigen) at the subspecies level using a MALDI-TOF MS platform with high specificity and sensitivity.
The results from the comparison of MALDI-TOF MS and serotyping show that the MALDI-TOF MS method is an accurate and sensitive approach for E. coli H antigen identification and typing. Reference strains representing all 53 E. coli H types were identified correctly by MALDI-TOF MS. Of 85 clinical isolates tested by MALDI-TOF MS-H, 75 identified MS-H types (88.2%) matched results obtained from traditional serotyping. This new method had a much shorter turnaround time, higher throughput, and lower cost than serotyping. |
 |
 |
 |
 |
|
Mascot tip of the month
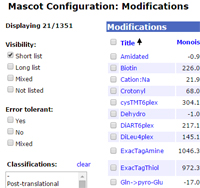
Mascot takes its list of modifications from the Unimod database, which now contains 1351 entries. In the past, if you wanted to change which entries appeared in the short list of common modifications, or which entries were considered in an error tolerant search, the procedure was somewhat painful, and your changes were lost if you updated your local unimod.xml file by downloading a later one.
The modifications module of the configuration editor has been much improved in Mascot Server 2.5. Now, you can easily select a group of modifications to be moved in to or out of the short list or the list used in an error tolerant search. Best of all, your changes are remembered when unimod.xml is updated. This includes which list a modification appears on, new entries, and changes to existing Unimod entries.
If you are updating to Mascot Server 2.5, please see the note describing how to migrate your old changes on the on the technical support page.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by AB Sciex, Agilent, Bruker, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|