|
To view this email as a web page, click here. |
 |
|
Welcome
As the holiday season approaches, we'd like to make a slight departure from proteomics and share some of the interesting history of our location on Baker Street, London.
This month's highlighted publication looks into the role of methionine oxidation in the oxidative stress response of Arabidopsis.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how you can share your search and quantitation results with colleagues by using Mascot Distiller as a free project viewer.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Baker Street
From Sherlock Holmes to the Secret Service to the Beatles, our street in London has been involved in a range of interesting cultural and historical events.
Read about Baker Street before Matrix Science settled at number 64. Warning: this article contains no proteomics! |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
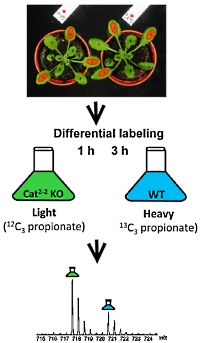
Protein Methionine Sulfoxide Dynamics in Arabidopsis thaliana under Oxidative Stress
Silke Jacques, Bart Ghesquiere, Pieter-Jan De Bock, Hans Demol, Khadija Wahn, Patrick Willems, Joris Messens, Frank Van Breusegem and Kris Gevaert
Molecular & Cellular Proteomics (2015) 14: 1217-1229
This work investigates methionine oxidation, which is a reversible post-translational modification and a mechanism by which proteins perceive oxidative stress. COFRADIC technology was used to specifically isolate peptides carrying in vivo oxidized methionine.
Arabidopsis catalase 2 knock-out and wild type plants were oxidized by increased concentrations of hydrogen peroxide. The proteins were extracted, desalted, and digested. The peptides were then fractionated and labelled. Those originating from cat-2 knockout were 12C3 propionated, whereas the wild type was 13C3 propionated. LC/MS/MS analysis and database searching resulted in the identification of 513 oxidation sites in 403 proteins.
Quantification of methionine oxidation using Mascot Distiller revealed 55 MetOx peptides in 51 proteins that were significantly more oxidized in the catalase knock-out. The data have been uploaded into the PRIDE database, project accession number PXD001286. |
 |
 |
 |
 |
|
Mascot tip of the month
An often over-looked feature of Mascot Distiller is that it can be installed on any Windows computer without a license and used as a project viewer.
The licensed copy of Distiller, used to create the project, should include the search toolbox so that search results can be imported and saved.
You need to make both the raw MS data file(s) and the Mascot Distiller rov file available to your colleague, but they do not need access to the Mascot Server as everything is stored in the rov file. Your colleague will be able to browse the raw data and examine both search and quantitation results, off-line. This is particularly useful for core labs distributing results involving quantitation or when people want to inspect matches that may be evidence for PTM's |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|