|
To view this email as a web page, click here. |
 |
|
Welcome
ASMS is fast approaching, and we would like to invite you to our breakfast meetings on June 6th and 7th in San Antonio.
In this month's featured publication the authors have developed a method for deeper analysis and increased coverage of histones using an Orbitrap.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains what to do if your search mysteriously crashes after 1 day.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Join Us at ASMS in San Antonio
We would like to invite you to join us for breakfast
at the upcoming ASMS meeting in San Antonio in June. We are hosting two sessions where you can learn about the latest applications and developments
in database search and quantitation. There is no charge for attending these meetings, but advance registration is required.
Monday 6 June, 7:00 am - 8:00 am
Ancient proteomes - proteomics in archaeology, palaeontology and forensics
presented by Michael Buckley, Manchester University
Mass tolerant and error tolerant searches: how do they compare?
presented by Matrix Science
Tuesday 7 June, 7:00 am - 8:00 am
From Samples to Biomarker Discovery: The keys for Large Scale Proteomics
presented by John Corthesy, Nestlé Institute of Health Sciences
New features in Mascot Distiller - MSE, MS3 reporter ion quant, and more
presented by Matrix Science
CLICK HERE FOR DETAILS AND REGISTRATION |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Analyses of Histone Proteoforms Using Front-end Electron Transfer Dissociation-enabled Orbitrap Instruments
Lissa C. Anderson, Kelly R. Karch, Scott A. Ugrin, Mariel Coradin, A. Michelle English, Simone Sidoli, Jeffrey Shabanowitz, Benjamin A. Garcia and Donald F. Hunt
Molecular & Cellular Proteomics (2016), 15, 975-988
Histones are the main protein component of chromatin and play a major role in the regulation of gene expression through complex patterns of post-translational modifications, histone variants, and proteolysis. To overcome some of the current limitations in the MS characterization of these histones, the authors have developed a new approach using a modified Orbitrap instrument.
They were able to greatly increase the fragment ion current by putting a reagent ion source in the front of the instrument, and using the C-trap for the storage of fragments from multiple iterations of ion/ion reactions. After sufficient accumulation, the ions were then transferred to the Orbitrap for measurement. This improves signal to noise and enables the acquisition of good quality spectra on a time scale more compatible with LC.
Histones from butyrate-treated HeLa cells were analyzed by top down LC/MS/MS using ETD. Several unique histone proteoforms were identified with up to 81% sequence coverage, and with a variety of modifications such as single amino acid substitutions, methylations, and acetylations. Additionally, truncated forms of histones H2A and H2B were characterized with up to 93% sequence coverage. |
 |
 |
 |
 |
|
Mascot tip of the month
From time to time, a Mascot Server user will report that some of their searches are crashing. When we try to spot a pattern, it becomes clear that the searches fail after approximately 86,400 seconds, a number you may recognise as corresponding to 1 day. The searches are being killed by IIS, the Windows web server. When Mascot is installed, the timeout for Mascot CGI processes is increased from the default of 900 seconds to 1 day, which is more than enough in most cases ... but not all.
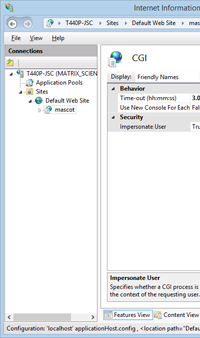
If your searches might need more time, and you are using IIS7 or later, you can easily increase this timeout. From the Windows Start menu, go to Control panel, Administrative Tools, Internet Information Services (IIS) Manager. On the connections tree, expand Sites and Default web site and select Mascot. In the central pane, double click the CGI properties icon. The CGI time-out will be displayed and can be edited. Note that the format to specify a full day is 1.00:00:00. If you make changes, choose Apply in the Action pane.
For IIS6 (XP and Server 2003), you have to use a script to make this change. The procedure is described in Appendix D of the Mascot Installation and Setup manual (follow the link on your local Mascot home page). |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|