|
To view this email as a web page, click here. |
 |
|
Welcome
We enjoyed seeing many of you at the ASMS conference in San Antonio. If you weren't able to attend our breakfast meetings,
PDFs of the presentations are now online.
We highlight some important developments in the new release of Mascot Distiller.
In this month's featured publication, the authors use a quantitative approach to evaluate the circadian rhythm of proteins in mice.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains why searches may appear to crash on a heavily loaded Windows server.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Mascot Distiller 2.6 Now Available
Earlier this month, we released a new version of Mascot Distiller,
a versatile tool set that is complementary to Mascot Server.
Distiller reads native file formats from all vendors, performs peak picking, Mascot search submission, and intensity-based quantitation for a variety of label-free or labeled workflows.
Enhancements in the new version include:
- Support for Waters MS^E data, including quantitation
- Reporter ion quantitation (iTRAQ and TMT) using MS3scans
- Summing or merging scans together according to scan type
- Sciex ClearCore library for much faster peak picking of Wiff files.
- Searching peaklists using different ions series depending on the scan type
- Entirely revised set of default processing options files
Read more about these new capabilities,
and try Mascot Distiller with your own data by taking advantage of our
free 30 day evaluation. |
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
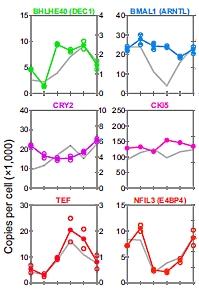
Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins
Ryohei Narumi, Yoshihiro Shimizu, Maki Ukai-Tadenuma, Koji L. Ode, Genki N. Kanda, Yuta Shinohara, Aya Sato, Katsuhiko Matsumoto, and Hiroki R. Ueda
PNAS (2016) 113, E3461, online: May 31, 2016
This study looked at the core circadian clock proteins in a mammalian system and quantified them with multiplexed SRM-based targeted proteomics. The authors used LC/MS-based "quantification by isotope labeled cell-free products" (MS-QBiC). This method uses a reconstituted cell-free protein synthesis system for high-throughput preparation of internal standards.
After 2 weeks of light/dark conditioning of the mice, livers were collected and analyzed at 6 time points using 120 QBiC peptides. Sixteen proteins from 20 selected circadian clock proteins were successfully quantified from the mouse livers over a 24-h time series, and 14 proteins had circadian variations. The study also demonstrated that single-time-points can predict body time, showing that expression levels of each circadian protein are potential markers for body time detection in human samples. |
 |
 |
 |
 |
|
Mascot tip of the month
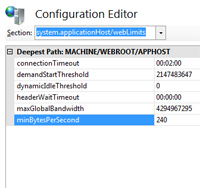
Under heavy system load, Mascot searches on a Windows server can appear to crash randomly. Repeating the same search at a quieter time succeeds. The IIS log contains Timer_MinBytesPerSecond error messages.
The problem is caused by communication between server and client becoming very slow. Recent versions of IIS set a minimum of 240 bytes / sec.
If the communication rate drops below this threshold, IIS kills the search.
For IIS 7.0 and later (Vista onwards) you can disable this threshold from the Windows control panel:
- IIS Manager
- Select Server
- Configuration editor
- Section: system.applicationHost/webLimits
- Set MinBytesPerSecond to 0
- Apply and re-start IIS
For IIS 6.0 (Windows Server 2003 and Windows XP Pro x64) the setting is MinFileBytesPerSec. KB919797 describes how to disable it. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|