|
To view this email as a web page, click here. |
 |
|
Welcome
Our lead article describes how Mascot security can be used to control access to a shared Mascot Server as well as
allow one user group to run bigger or longer or more complex searches than another.
Five modifications in the Dark Matter Challenge have been explained. Fifteen still to puzzle over.
Click here to apply your chemistry expertise.
This month's highlighted publication describes a new software tool to elucidate protein N-termini and quantify acetylation.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns quantitation of extreme ratios.
Please have a read and feel free to contact us if you have any comments or questions. |
|

|
|
 |
 |
 |
|
Stay in control with Mascot security
If multiple research groups within an organisation are using Mascot, it is usually more efficient and cost-effective to share
a single, large server than have multiple small servers. Initially, there may be some reluctance to share a server; possible privacy issues or concerns about someone hogging machine time so that your own searches are delayed or run slowly. Mascot Security provides a solution on both counts.
Regarding privacy, Mascot security provides mechanisms to limit access to result reports, databases, and even custom modifications.
Specific examples of how Mascot security can be configured to ensure privacy are described
in this article.
Regarding priority, you can set the maximum priority for each group to ensure searches from certain groups get a greater share of processor time.
Users can decrease the priority for their non-urgent jobs, but they cannot increase it above this maximum.
Novices and optimists tend to submit searches against the largest database they can find with many variable
modifications and no taxonomy filter; searches that seem to take forever.
You can counter this by limiting the availability of very large databases, such as nr, restrict groups from performing 'no enzyme' searches, and limit the number of variable modifications that can be selected.
Full details of priority related security settings can be found here.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
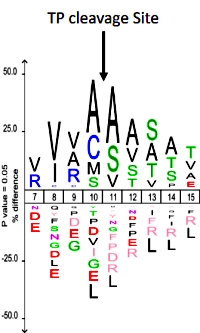
EnCOUNTer: a parsing tool to uncover the mature N-terminus of organelle-targeted proteins in complex samples
Willy Vincent Bienvenut, Jean-Pierre Scarpelli, Johan Dumestier, Thierry Meinnel and Carmela Giglione
BMC Bioinformatics, 2017, 18:182
The authors have developed a new tool called EnCOUNTer (Extraction and Calculation Of Unbiased N-Termini) which can determine the position of the protein N-terminus as well as the N-terminal acetylation yield for hundreds to thousands of candidates.
The workflow consists of acetylated and digested proteins fractionated on an SCX column, then analyzed by LC/MS/MS. Peptide N-terminal acetylation status was investigated using the Mascot Distiller quantification option, looking at the d3-Acetyl (chemically induced modification) or d0-Acetyl (endogenous modification). The EnCOUNTer tool requires the Mascot Distiller exported file, the associated Mascot search results and an optimized parameter file.
The N-terminal scoring was delineated according to six coefficients: i) peptide starting position, ii) residues around the starting position, iii) characterized N-terminal modifications, iv) alternative start positions at the vicinity of the starting position, v) matched peptide redundancies and iv) the Localization score. When applied to A. thaliana cell lysate, the EnCOUNTer tool was able to characterize more than 1200 N-termini of which almost 600 were quantified for N-terminal acetylation yield.
|
 |
 |
 |
 |
|
Mascot tip of the month
If you are performing quantitation using SILAC, or any other isotopic label that includes only a small number of heavy atoms, remember that labelling is never 100% complete. This means that ratios can be very inaccurate when the 'heavy' protein is strongly up-regulated relative to the 'light'.
For example, imagine sample A is labelled and sample B is unlabelled and labelling is 99% complete. If the true A/B ratio for a peptide is 1000, the measured ratio would be 90 because sample A is 1% unlabelled. An error in the ratio of more than an order of magnitude.
In most cases, this doesn't matter. If it does, you may need to perform technical replicates with the labelling inverted, and take values for extreme ratios from the experiment where the up-regulated peptide is unlabelled. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|