|
To view this email as a web page, click here. |
 |
|
Welcome
Mascot Server supports 17 different ion series. We use EThcD to illustrate which ones to choose for a new instrument.
This month's highlighted publication is a novel method for improved peptide identification and quantitation using 12C labelling.
(Yes, that's not a typo!)
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to modify the Mascot Server taxonomy tree.
Please have a read and feel free to contact us if you have any comments or questions. |
|

|
|
 |
 |
 |
|
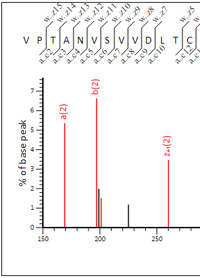
17 Ion series - which to choose?
Though there are 17 ion series available for instrument definitions in Mascot Server, it is best
not to select them all. Including series that are not present in the data reduces discrimination and sensitivity
because it increases the opportunities for false matches.
To look at the effect of including too many ion series, we examined a set of Electron-Transfer/Higher-Energy Collision Dissociation data. EThcD produces a combination of b/y and c/z fragment ions in a single spectrum, often improving identifications and site localisation for modifications. The data was from lysed HeLa cells digested with trypsin, peak picked using Mascot Distiller and searched using these settings: Trypsin/P with 2 missed cleavages, Carbamidomethyl (C) fixed, Oxidation (M) variable, peptide tolerance 15 ppm, fragment tolerance 30 ppm.
The difference in PSM count at 1% FDR considering all ion series versus an optimised selection for EThcD is only ~1%. Mainly because this is high accuracy data being searched with narrow mass tolerances, so the presence of unnecessary ion series doesn't lead to large numbers of false fragment ion matches. If we search with wider mass tolerances, 0.5 Da for both peptide and fragments, the effect becomes stronger, with a difference in PSM count of ~6%. Go here to read more.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
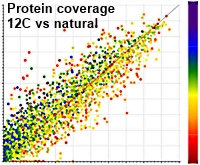
A Simple Light Isotope Metabolic labeling (SLIM-labeling) strategy: a powerful tool to address the dynamics of proteome variations in vivo
Thibaut Leger, Camille Garcia, Laetitia Collomb and Jean-Michel Camadro
Molecular & Cellular Proteomics, in press, published August 18, 2017
The authors have developed a new approach to address some of the limitations imposed by current quantitative proteomics methods. With metabolic labelling or stable isotope reagents, there are challenges due to the variable mass shifts or the ion suppression from additional peptides present causing lower identification efficiencies.
The paper describes bottom-up proteomic analyses of the pathogenic yeast, C. albicans, grown on a synthetic medium containing either normal glucose or depleted glucose containing only 12C carbon atoms as the sole carbon source. The use of 12C resulted in an increase in the intensity of the monoisotopic ion, markedly improving bottom-up proteomics analyses.
12C incorporation resulted in better overall scores per identified peptide with average scores up to 28% higher leading to an average increase of the protein identification scores of 36%. The number of identified peptides and proteins in C. albicans also increased by 14% and 11%, respectively, when applying a 1% FDR filter after 12C enrichment in vivo.
|
 |
 |
 |
 |
|
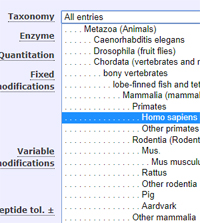
Mascot tip of the month
The taxonomy tree that appears in the search form and the protein family summary is fully configurable.
You can add or remove individual taxons, use names you prefer (cattle or cow or beef or bovine or bos taurus?), and create
nodes for specific experiments, e.g. human + mosquito.
There isn't a module for this in the configuration editor. You have to open the file
(mascot/config/taxonomy) in a text editor.
Each node is defined by a block of four lines. The first line of each block starts
with the keyword Title:, followed by the text to be displayed in the drop-down list.
Next, a line starting with the keyword Include:,
followed by one or more NCBI taxonomy IDs separated with commas. The best way to
look-up taxonomy IDs is to use the NCBI taxonomy browser.
The third line starts with the keyword Exclude:, and can be used to filter out
taxonomy IDs. The final line contains just an asterisk. For more complete details,
refer to chapter 9 of the Mascot Server Installation and Setup manual, which
is available via a link on your local Mascot Server home page. |
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|