|
To view this email as a web page, click here. |
 |
|
Welcome
There are compelling reasons to reconsider your alkylation strategy if you are using iodoacetamide or iodoacetic acid.
This month's highlighted publication shows the importance of detergent selection for characterizing membrane protein complexes.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is a reminder of how you can use the Database Status pages to change the priority of a running search, or even kill it!
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Alkylate with Caution
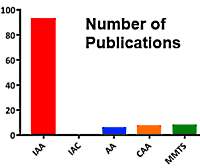
Iodine-containing reagents can cause very extensive over-alkylation, especially for in-gel digested samples where the stoichiometry is not well controlled. Unfortunately, according to the literature, iodoacetamide is easily the most widely used reagent.
If only carbamidomethyl (C) is selected for the search, large numbers of PSMs are likely to be missed. Trying to remedy this by including additional modifications increases the search space so that sensitivity is reduced, searches take longer, and the signal to noise suffers because the signal for any particular peptide is spread over multiple precursors. When it comes to quantitation, the latter effect can be a major source of error.
There is no need to tolerate these problems. Simply switch to an alkylation reagent that does not contain iodine. Go here to read more.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Impact of Detergents on Membrane Protein Complex Isolation
Yu-Chen Lee, Jenny Arnling Baath, Ryan M. Bastle, Sonali Bhattacharjee, Mary Jo Cantoria, Mark Dornan, Enrique Gamero-Estevez, Lenzie Ford, Lenka Halova, Jennifer Kernan, Charlotte Kurten, Siran Li, Jerahme Martinez, Nalani Sachan, Medoune Sarr, Xiwei Shan, Nandhitha Subramanian, Keith Rivera, Darryl Pappin, and Sue-Hwa Lin
Journal of Proteome Research, published online November 7, 2017
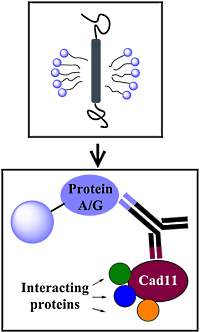
Detergents play an important role in the isolation of membrane protein complexes, affecting the native folding of the proteins, their binding to antibodies, and their interaction with partner proteins. In this paper, the authors investigated the effect of several commonly used detergents on Cad11 protein complex solubilization and coimmunoprecipitation.
Antibody pull-downs of the complex, coupled with iTRAQ quantitation, were used to determine the associating proteins. Seven detergents were examined and the authors found variations in the association of certain proteins with Cad11 depending on the detergents used.
DDM and Triton X-100 were found to be more efficient in solubilization and immunoprecipitation of Cad11 and provided 40 new candidate Cad11-interacting proteins. The authors suggest that using a panel of detergents may lead to a more comprehensive view of the composition of complexes.
|
 |
 |
 |
 |
|
Mascot tip of the month
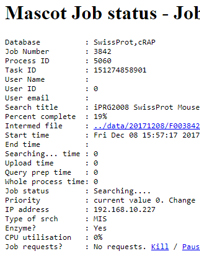
The Database Status page is where you can find status information for both databases and running searches on your Mascot Server. The links at the top of the status page to the search log, error log, and monitor log are fairly obvious. It may not be so clear that you can also control running searches.
Imagine you have started a lengthy search of a large database, such as NCBIprot, and you now want to submit some small searches where the results are needed very urgently. On the Database Status page, click on the NCBIprot database name to get a list of running searches. If the database has been updated since Mascot Monitor was started, there will be two entries, one for the old file and one for the new, so make sure you choose the one labelled 'In use'.
Locate the lengthy search from the submission time or some meta data such as the search title and click on the job number to get detailed information. You'll see links to reduce the priority, so that it gets a smaller share of CPU time. Priority is measured in negative nice units, with -20 being lowest possible priority through to +20 for highest.
There is also a link to pause the search, but don't forget to come back later and resume it. If you decide that you no longer want the search to run at all, there is a link to kill the search. More about this and similar topics in module L of the webcast training course.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|