|
To view this email as a web page, click here. |
 |
|
Welcome
If you are attending the ASMS Annual Conference in San Diego next month, please stop by and visit us at Booth #522. You are also invited to join us at our ASMS breakfast meetings on June 4th and 5th in the Convention Center.
To mark the 20th anniversary of Matrix Science, we are offering a 20% discount on Mascot Server version updates.
We look at the implications of key sequence database resources becoming HTTPS-only. In connection with this, there is a new patch release for Mascot Server.
This month's highlighted publication describes a deep proteome analysis for the elucidation of citrullination sites in human tissues.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Matrix Science Breakfast Workshops at ASMS
Please join us at our annual User Meetings at the upcoming ASMS Conference in San Diego, California. These breakfast workshops take place on June 4th and 5th at the San Diego Convention Center, where we will describe the latest tools and techniques for database searching. We also have two featured speakers presenting their exciting applications.
Monday 4 June, 7:00 am - 8:00 am
Expanding the landscape of human phosphorylation
presented by Claire Eyers, Centre for Proteome Research, U. Liverpool
Mascot Distiller gets up to speed
presented by Matrix Science
Tuesday 5 June, 7:00 am - 8:00 am
Identification of Iodine-containing Alkylation Reagent related Artifacts and Polymer Contaminations in Proteomics Samples
presented by Dominic Winter, Institute for Biochemistry and Molecular Biology, U. Bonn
Using spectral libraries to pick a peck of pickled peptides
presented by Matrix Science
Please go here to register and reserve your spot as seating is limited. Breakfast will be served. |
 |
 |
 |
 |
|
Stepping up security
An increasing number of web sites are dropping support for HTTP and moving to HTTPS, exclusively, because this provides a more secure mode of communication. This includes the sites of some of the organisations that provide resources and databases for proteomics research.
In late 2016, NCBI dropped support for HTTP requests and restricted their web resources to HTTPS. EBI went HTTPS by default in October 2017 and UniProt has announced that it will go HTTPS-only in June 2018.
An HTTP link in a web page will continue to work because all popular web browsers automatically switch to HTTPS. On the other hand, the full annotation text embedded in a Mascot Protein View report is fetched by a script from an external URL. Versions of Mascot Server prior to 2.6 assumed these external calls would always use port 80. If the annotation text is only available via an HTTPS call, it will be missing from the report. Currently, this only affects NCBI databases, but it will also apply to EBI and UniProt if and when they turn off HTTP.
If a sequence database file is only available via HTTPS, current releases of Database Manager will not be able to download it. This isn’t a problem right now because the NCBI, NIST, and EBI file repositories use FTP protocol. It will become a problem for Fasta files created using a query, such as UniProt proteomes, when HTTP is not available. To address this, we have released a patch for Mascot Server, which is described below
Please go here to read about these issues and suggested work-arounds in greater detail.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
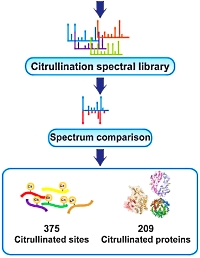
Mining the human tissue proteome for protein citrullination
Chien-Yun Lee, Dongxue Wang, Mathias Wilhelm, Daniel Paul Zolg, Tobias Schmidt, Karsten Schnatbaum, Ulf Reimer, Fredrik Ponten, Mathias Uhlen, Hannes Hahne and Bernhard Kuster
Molecular & Cellular Proteomics, Published online April 2, 2018
Citrullination is the conversion of arginine to citrulline by peptidylarginine deiminases leading to the mass increment of 0.9840 Da. This mass is identical to the common deamidation of Asn/Gln residues, leading to ambiguity in database searching of data from complex digests.
Citrullination has been implicated in many cellular processes such as terminal epidermal differentiation, apoptosis, central nervous system (CNS) stability, immune response, gene regulation, and embryonic development.
To address the sparsity of information on specific citrullination sites, the authors mined deep proteomes of 30 human tissues for endogenous protein citrullination. To analyze the ~70 million MS/MS spectra, generated by 1,116 LC-MS/MS runs, they used the neutral loss of isocyanic acid, the immonium ion of citrulline, manual spectrum interpretation, as well as reference spectra of synthetic peptides. This resulted in the identification of 375 citrullinated sites on 209 human proteins across 26 human tissues.
|
 |
 |
 |
 |
|
Mascot Server 2.6.2
We are pleased to announce a patch release of Mascot Server. This adds HTTPS support to Database Manager, enabling files that are only available via HTTPS to be downloaded. Other bug fixes include support for hardware with more than 64 logical processors and a fix for an occasional cache file error in Protein View.
This free update for version 2.6 can be downloaded from the Mascot Server support page. If you have an earlier version of Mascot Server, this is a great time to buy an update because we are offering 20% off all Mascot Server updates to mark the 20th anniversary of Matrix Science. This limited-time offer is for orders received by 31st July 2018.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|