|
To view this email as a web page, click here. |
 |
|
Welcome
We enjoyed speaking with many of you at the ASMS meeting in San Diego earlier this month. If you didn't get a chance to attend our workshops, the company presentations are available here.
We describe how to use Mascot Database Manager to automate updates of genomic DNA databases.
This month's highlighted publication shows a method for quantifying the extent of phosphorylation using parallel reaction monitoring.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns troubleshooting a hung task in Mascot Daemon.
If you are running an old version of Mascot, time is running out to take advantage of our 20th anniversary offer of 20% discount on Mascot Server version updates.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Keeping genome databases up to date
Updating most databases in Mascot Server Database Manager is as simple as clicking on a button or creating a regular update schedule.
Occasionally, you need additional processing steps after downloading a file. For example, if you want to search an assembled genome,
we recommend splitting chromosome length sequences into shorter, overlapping segments.
It is possible to automate the complete procedure by creating a simple script to download the file, execute the splitter utility,
then prompt Database manager to copy the output files to the 'current' directory for the database.
This article includes a example script that downloads
and splits the Helicobacter pylori HPAG1 genome from the European Nucleotide Archive.
The script can also be used as a template for other types of processing, such as generating a Fasta file from sequences stored in a relational database.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Determination of Site-Specific Phosphorylation Ratios in Proteins with Targeted Mass Spectrometry
Lennard J. M. Dekker, Lona Zeneyedpour, Sandor Snoeijers, Jos Joore, Sieger Leenstra, and Theo M. Luider
J. Proteome Res. 2018, 17, 1654-1663
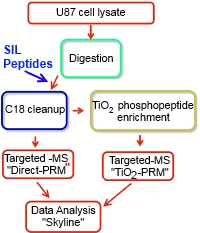
The authors developed a method to absolutely quantify phosphopeptides and to determine their percentage
of phosphorylation using parallel reaction monitoring (PRM) in combination with stable-isotope-labeled (SIL) peptides.
First a phosphoproteome profile is created by performing a data-dependent LC-MS measurement on enzymatic digested glioblastoma multiforme cell lysates that are enriched by TiO2. Second, a number of phosphopeptides were selected from this data set and used for a PRM approach with SIL peptides. In this PRM approach phosphorylated peptides as well as the nonphosphorylated counterparts including their SIL standards are targeted.
This resulted in the identification of 7774 unique phosphopeptides and 1980 pairs where the non-phosphorylated peptide partners were also observed. From this list, six peptide pairs were chosen based on their relevance to pathways that are known to be affected in glioma. This validation study showed that a reproducible quantity of phosphorylation could be determined with CV's of 6-13%.
|
 |
 |
 |
 |
|
Mascot Tip
If Mascot Daemon encounters an unexpected error, it is possible that a task may hang, with the task icon on the Daemon status tab displaying as an hour glass. If this happens to you, here are some things to try:
- First, check that your Mascot Server is operating correctly. Go to the database status page and check that the database you are trying to search is 'In use'. Make sure there aren't large numbers of searches running which have made the server unresponsive.
- If there are any searches running, click on the database name then on the job number to see the search title and other details. If a search submitted from your instance of Daemon is running, best to let it complete before trying to re-start Daemon. Otherwise, you risk Daemon re-submitting it.
- Mascot Daemon 2.5 and later: Exit the Daemon desktop application then stop the Daemon engine. This may be running in the system tray (right click the icon and choose exit) or as a Windows service (Windows control panel; Administrative tools; Services; Mascot Daemon Engine; Select; Stop). In Windows task manager, verify there is no running process called MascotDaemon.exe or MascotDaemonEngine.exe
Mascot Daemon 2.4 and earlier: Exit the Daemon desktop application then stop the Daemon service. This may be running in the system tray (right click the icon and choose exit) or as a Windows service (Windows control panel; Administrative tools; Services; Mascot Daemon Service; Select; Stop). In Windows task manager, verify there is no running process called Daemon.exe or DaemonService.exe
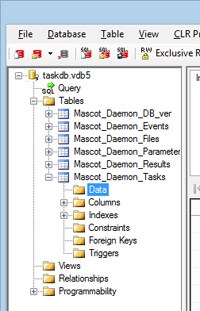
- Start Daemon. If the task is still hung, make a note of the task number. If the database engine is VistaDB, refer to the Daemon help (F1) for details of how to edit the task database. For other database engines, use whatever utility is available to manipulate the tables or execute SQL commands.
- Exit both the desktop application and the engine / service, as above. Delete the hung task from the Mascot_Daemon_Tasks table with an SQL command such as this, where task_UID is set to the problem task number:
DELETE FROM Mascot_Daemon_Tasks WHERE task_UID=502;
An alternative is to revert to an empty task database. For details, email support@matrixscience.com describing the problem and specifying the Daemon version and task database engine.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|