|
To view this email as a web page, click here. |
 |
|
Welcome
There are a few recurring misconceptions about how protein scores should be used.
This month's highlighted publication shows a new approach to identify and quantify the challenging persulfidation modification.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is essential reading if you are still using the Access database engine with Mascot Daemon.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Common myths about protein scores
Mascot Server is used in many different application areas by both mass spectrometry experts and non-experts. Over the years, we've spotted a few recurring misconceptions about how protein scores should be interpreted and used.
In particular, protein scores in MS/MS searches are only used to rank protein hits with the goal of putting proteins with lots of strong peptide evidence at the top of the list. Unfortunately, you sometimes see MS/MS papers with phrases like "all proteins identified with a Mascot score higher than 60 were considered reliable" or "peptides and proteins with a Mascot score higher than 35 and 50, respectively, were automatically accepted.". A protein score of 60 could mean the protein has one significant peptide match with score 60, or the protein could have 47 peptide matches each with score 14 and threshold 13.
Unsafe assumptions can also arise when trying to filter out false positive proteins. It is well known that accepting a protein on the basis of a single peptide match is very risky; these are so-called 'one-hit wonders'. Alarm bells should ring if you see statements such as "identified proteins exhibited at least one peptide with an individual Mascot score of p<0.05" or "proteins were accepted if they had at least one 'rank 1' peptide with a peptide ions score of more than 50.0."
Go here to read more details about the best use of protein scores and some pitfalls to avoid.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
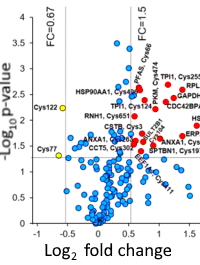
Site-Specific Quantification of Persulfidome by Combining an Isotope-Coded Affinity Tag with Strong Cation-Exchange-Based Fractionation
Qiong Wu, Baofeng Zhao, Yejing Weng, Yichu Shan, Xiao Li, Yechen Hu, Zhen Liang, Huiming Yuan, Lihua Zhang, Yukui Zhang
Analytical Chemistry 91 14860-14864 (2019)
The authors have developed a new approach to profile the challenging persulfidation modification. Previous methods have had some limitations in terms of false positives as well as underreporting, due in large part to the similar reactivity of the persulfide groups, disulfide bonds, and thiol groups.
The workflow entails first labeling persulfidated proteins with cleavable isotope-coded affinity tag (c-ICAT) reagents. After digestion, the labeled persulfidated peptides were selectively enriched with streptavidin beads and fractionated by strong cation exchange chromatography, followed by LC−MS/MS identification.
After confirming the efficiency of the method, they applied it to the site-specific identification of endogenous protein persulfidation from A549 cells. They identified 226 endogenous persulfidated sites on 160 proteins with 74% of the persulfidation sites newly discovered. Of these proteins, 71% were singly modified, 21% had 2 sites of persulfidation, while 2.5% had 4 or more.
|
 |
 |
 |
 |
|
Mascot Tip
The default database engine for Mascot Daemon changed from Access to VistaDB back in 2014 (Mascot Daemon version 2.5). If you are still using Access, please be aware of a serious problem caused by the November 2019 security update for Office 2010, 2013, and 2016. A Microsoft support document explains how to uninstall the update.
Our recommendation is to take this opportunity to switch to a different engine. If the task database is only used by a single instance Daemon, switching to VistaDB is the simplest option. If the task database is shared, switch to PostgreSQL, SQL Server, or MySQL. The Mascot Daemon Backup and Restore facility makes it easy to copy the database contents from the old system to the new; more details in the Daemon help under Getting Started; Database Engines. (Thanks to Mike Naldrett of U. Nebraska-Lincoln, who was the first to report this problem and identify the cause.)
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|