|
To view this email as a web page, click here. |
 |
|
Happy New Year!
A new release of Mascot Server will soon be available and we have a preview below.
This month's highlighted publication examines collision energy dependence of fragmentation and optimal strategies for best coverage of monoclonal antibodies.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes a change in the way a Uniprot proteome is specified for download.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Coming attractions:
Mascot Server 2.7
Mascot Server 2.7 is now running on our website and should be ready for release later this month. We have a number of new and enhanced features that many of you have requested, and here is a sampling.
- Crosslinked peptides Pairs of peptides linked by intact crosslinks can now be matched in a Mascot search by selecting a named crosslinking method.
- Protein FDR Search results for auto-decoy searches now report a Protein false discovery rate estimated using a MAYU type of approach.
- Non-standard modification names A new configuration file can be used to map modification names used in a spectral library to their Unimod equivalents.
- Quantitation Summary A CSV Quantitation Summary, in which the rows correspond to proteins and the columns contain expression data for multiple samples, can be created in Mascot Daemon.
- Improved limits on variable modifications Better site localization by introducing separate caps on the number of distinct modifications in a PSM, the number of modified sites in a PSM, and the total number of arrangements to be tested.
Go here to read more details about the upcoming release of 2.7, or contact us to request a quotation for an update.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
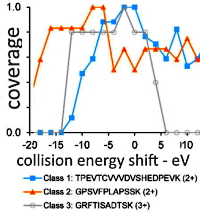
Optimal Collision Energies and Bioinformatics Tools for Efficient Bottom-up Sequence Validation of Monoclonal Antibodies
Agnes Revesz, Tibor Andras Rokob, Dany Jeanne Dit Fouque, Daniel Huse, Viktor Hada, Lilla Turiak, Antony Memboeuf, Karoly Vekey and Laszlo Drahos
Analytical Chemistry 91 13128-35 (2019)
The authors have developed software called Serac capable of automatically computing and reporting protein "confirmed sequence coverage" (CSC) from LC-MS/MS runs that were processed via Mascot.
They analyzed two therapeutic mAb digests (rituximab and trastuzumab) and mapped the collision energy dependence of peptide and protein CSC to find the optimal settings. They also looked at increasing CSC by combining information from several sources (multiple spectra on different charge states, multiple runs collected under different settings) and at multiple levels (peptide, amino acid, fragment ion).
They proposed that an optimal workflow would include 2-3 LC-MS/MS runs (at collision energies 6-10ev apart) that are combined at the fragment ion level. This resulted in a confirmed sequence coverage 25%-30% above that from a reference approach involving a single run and no dedicated data processing.
|
 |
 |
 |
 |
|
Mascot Tip
Uniprot have dropped the complete proteome classification. If you have this keyword in an old download URL in Database Manager, you'll need to fix it before trying to update the files. Go to the Uniprot Proteomes site and search by taxonomy ID or keyword. The proteome ID is the correct identifier to use in the Fasta download URL.
Keywords for well studied organisms will return a number of potential matches, so be careful to choose the correct one for your samples. Entering "mouse", for example, returns 13 reference proteomes, but most of these are for unrelated organisms that happen to include this word in the description. Uniprot have added two columns of information to help judge how complete a proteome might be, and protein count is another good indicator.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|