|
To view this email as a web page, click here. |
 |
|
Welcome
We have improved the handling of variable modifications in Mascot Server, enabling optimization for speed or depth.
This month's highlighted publication describes the discovery of a specific modification that causes a protein to induce proteome-wide dysfunction.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month is a reminder about support and updates for Mascot Server.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Variable modifications - searching with depth or speed
We've made improvements in Mascot Server 2.7 so that you now have greater flexibility to control how variable modifications are handled. New parameters are available to provide increased speed or more depth of coverage, according to your samples, data, and the questions being asked.
The three parameters have default values to provide results similar to Mascot 2.6 and earlier, but you can adjust the values to suit your experiments:
- Maximum number of different variable modifications per peptide
- Maximum number of modified residues per peptide
- Maximum number of arrangements of an individual set of variable modifications
We have two illustrations of the effect of these parameters. The first is an error-tolerant search for a lightly modified sample, a subset of a mouse label-free dataset with 20,000 MS/MS spectra. Using tighter values for these modification settings, we were able to reduce the search time by 35% while maintaining the results, as compared with Mascot 2.6.
For the second case we used a middle-down human histone H4 dataset, where the study was looking at changes in modification patterns across the cell cycle on the N-terminal 23 residues. Adjusting the parameters allowed us to not only identify more proteoforms, but also to have more confident site localization.
Go here to read about this in detail.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
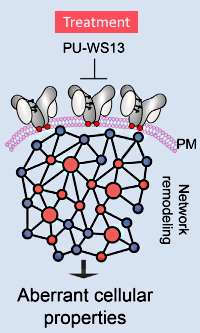
Molecular Stressors Engender Protein Connectivity Dysfunction through Aberrant N-Glycosylation of a Chaperone
Pengrong Yan, Hardik J. Patel, Sahil Sharma, Adriana Corben, Tai Wang, Palak Panchal, Chenghua Yang, Weilin Sun, Thais L. Araujo, Anna Rodina, Suhasini Joshi, Kenneth Robzyk, Srinivasa Gandu, Julie R. White, Elisa de Stanchina, Shanu Modi, Yelena Y. Janjigian, Elizabeth G. Hill, Bei Liu, Hediye Erdjument-Bromage, Thomas A. Neubert, Nanette L.S. Que, Zihai Li, Daniel T. Gewirth, Tony Taldone, and Gabriela Chiosis
Cell Reports 31(13), 107840, (June 30, 2020)
The authors employed a wide range of experimental techniques to investigate the structural and functional changes caused by stress-induced N-glycosylation of a chaperone protein, GRP94.
Using native and SDS-PAGE, siRNA knock-down, enzymatic deglycosylation, CRISPR/Cas9 mediated mutants and knock-outs, cell viability by flow cytometry, PK/PD/tox studies, and identification of N-glycosylation sites using LC-MS/MS, they demonstrated that the N-glycosylation transforms the GRP94 from a folding to a scaffolding protein.
This specific increase in N-glycosylation promotes a conformational state that allows for stable interactions with oncoproteins at the plasma membrane. Using a small-molecule purine compound, the authors also showed that they could selectively inhibit the formation of this aberrant form, indicating a potential therapeutic route.
|
 |
 |
 |
 |
|
Mascot Tip
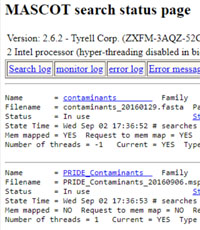
When warranty or annual support for a Mascot Server licence is due to expire, we email a renewal invitation to the registered contact. We suspect that quite a few of these reminders will have been overlooked in the chaos of the last few months. If you wish to check your support status, just email the product key displayed at the top of the Database Status page to support@matrixscience.com
The Database Status page also shows the software version. For licences that are under support or warranty, when there is a new release of Mascot Server, we automatically email an update key and download link. For 2.7, an email was sent to the registered contact on January 30th of this year. If you are running an earlier version, and suspect your update email has been overlooked, please contact support@matrixscience.com
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|