|
To view this email as a web page, click here. |
 |
|
Welcome
We look at validating matches to intact crosslinked peptides.
This month's highlighted publication shows a new method for separating and identifying protein N-terminal peptides.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains a quirk in the way clients sometimes display error messages.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Is your crosslinked peptide ID correct?
Chemical crosslinking is a powerful technique for structural elucidation, but the geometric increase in possible matches makes interpretation challenging. Whether it is a looplinked sequence, a linear sequence with monolink or several different peptide pairs (potentially also modified), Mascot 2.7 uses the same scoring system, so the scores for all matches to a query are directly comparable. This makes it straightforward to compare and rank the candidate matches.
Using recently published data obtained from a library of known synthetic DSS-crosslinked peptides, we assessed the true FDR for significant crosslinked matches. The search results show a good number of monolink matches and intact crosslinks in Cas9, including many strong matches. We also analyzed the various false positives in detail and identified several linear peptides with monolinks, as well as some implausible alpha-beta peptide combinations.
A helpful strategy for maximizing sensitivity while controlling for false positives is:
- Make sure the search space is covered
- Only include the necessary protein sequences in the search, no more
- Always look at the fragmentation and alternatives to the rank 1 match
Please go here to read the details.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
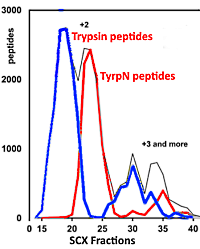
Isolation of acetylated and unmodified protein N-terminal peptides by strong cation exchange chromatographic separation of TrypN-digested peptides
Chih-Hsiang Chang, Hsin-Yi Chang, Juri Rappsilber and Yasushi Ishihama
Molecular & Cellular Proteomics November 2, 2020
The authors have developed a new approach to characterize the N-termini of proteins by enriching N-terminal peptides without the need for derivatization or complex processing. The method uses the protease TyrpN and then leverages strong cation exchange (SCX) chromatography to separate peptides based on the number of positive charges as well as the localization of the charges. This results in the separation of the protein N-terminal peptides from internal peptides.
They evaluated this separation by comparing the retention time in SCX for approximately 4,000 peptide pairs having sequences that differ only in the position of terminal Lys/Arg. The TrypN-digested peptides clearly showed a stronger SCX retention than the Typsin analogs. This is because the TrypN peptides carry two positively charged groups at the N-terminus, whereas the positive charge of the C-terminal Lys/Arg of trypsin peptides was partially neutralized by the α-carboxy group.
Applying this method to a human embryonic kidney cell lysate, they identified 1,550 acetylated and 200 unmodified protein N-terminal peptides in a single LC/MS/MS run with less than 3% contamination with internal peptides.
|
 |
 |
 |
 |
|
Mascot Tip
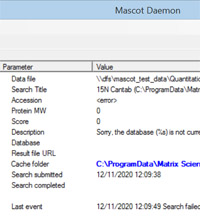
If you use a client such as Mascot Daemon or Proteome Discoverer to submit searches, you may have noticed that warnings or error messages from the Mascot Server are sometimes incomplete. For example, the Daemon status tab may report something like this:
Sorry, the database (%s) is not currently available for searching A bit cryptic. The '%s' is a placeholder for a string, and the message should have read
Sorry, the database (Uniprot_arabidopsis) is not currently available for searching If you ever see this on your system, and the complete message would make it easier to figure out what is causing the problem, you can always get the full text by looking on the Mascot Server. In a web browser, from your local Mascot home page, follow the link to Database Status, then Error log. It shouldn't be difficult to locate the complete message by matching up the date and time with that of the client event.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|