Unipept and Mascot
Drawing conclusions from protein-level data is complicated in environmental and metaproteomics studies, where the sample is a mixture of hundreds or thousands of proteomes. The Unipept database is a useful, complementary resource for interrogating metaproteomics data and can be used in conjunction with Mascot’s protein inference.
Human gut example
Identify proteins by more than ‘gut’ feeling discussed analysing a human gut sample with Mascot. The data were searched against the draft genomes of human gut microbial species as well as a matched metagenome constructed from next-gen sequencing data. The latter aims to fill the gaps in the search space where no genome is available.
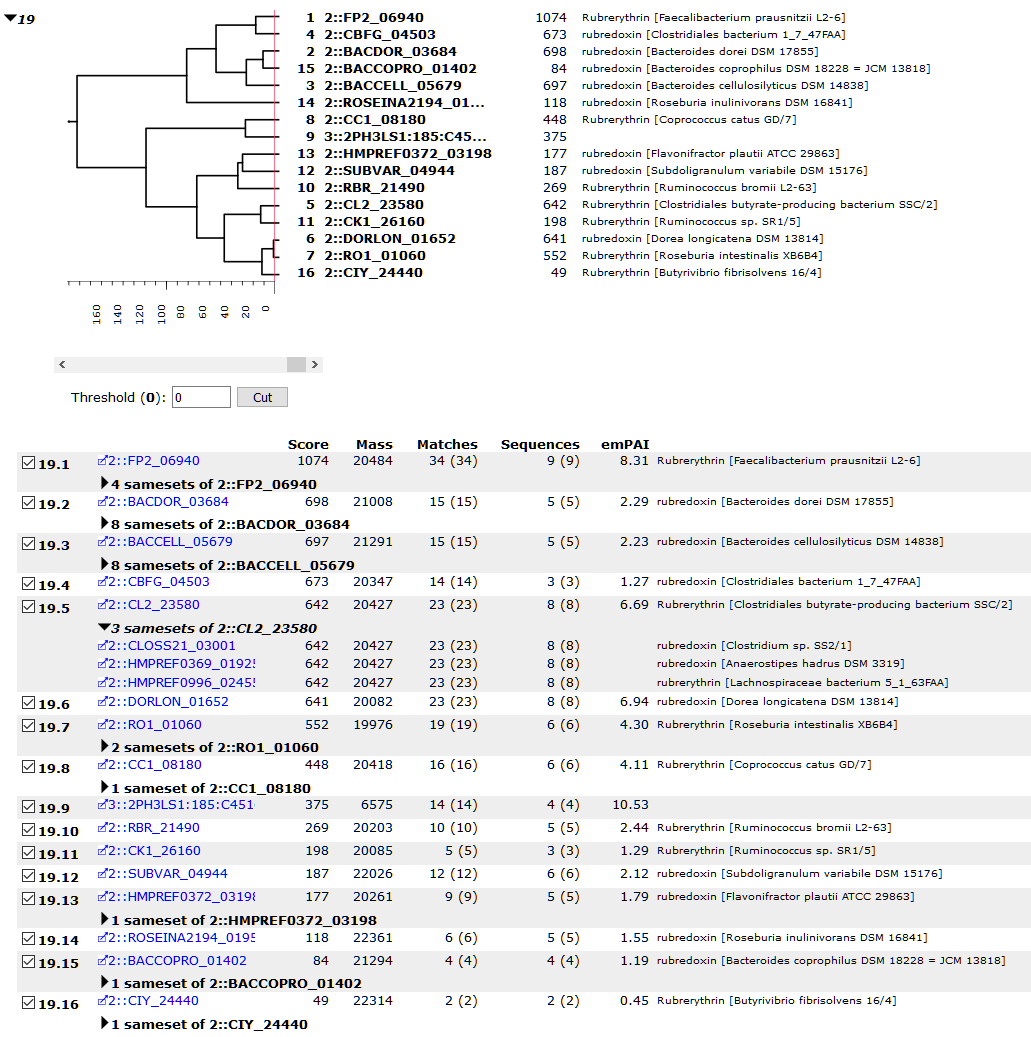
The Protein Family Summary report groups proteins by shared peptide evidence,
which is often correlated with protein sequence similarity.
Human proteins cluster in families separate from bacterial proteins.
Bacterial proteins similar to each other cluster in the same family
without taxonomic restriction, like rubredoxin and rubrerythrin in
family 19 in the earlier blog post (screenshot reproduced above).
Peptides found only in the matched metagenome either cluster with
proteins from the draft genomes or appear as one-hit wonders
.
Missing from Protein Family Summary is an overview of species or other kinds of taxonomic groupings. This is where Unipept comes in handy.
Unipept
Unipept is a web application with a browser interface. The database maps every tryptic peptide in UniProtKB to its lowest common ancestor (LCA). A peptide’s LCA could be a single species, like the Hymenobacter peptide VVSTNDANYR. Conversely, a peptide shared between many species across different phyla, like AELENTDSDYDR, is assigned to ‘Bacteria’. The recently released Unipept Desktop application is convenient for desktop use with larger data sets.
One way to use Unipept is importing all significant peptide sequences from the Mascot search. In Protein Family Summary, set PSM FDR to 1% and export the results as CSV. Untick Include same-set protein hits, set Include sub-set protein hits to zero and ensure Max. number of hits is AUTO. The CSV file contains all significant peptide matches. Open the file in Excel, sort the match table by pep_rank and delete any rows with pep_rank > 1. Then, sort by the pep_seq column and copy the peptide sequences into a text file. Import the text file in Unipept Desktop as a new assay (ensure duplicates are filtered out). A couple minutes later, the assay has been analysed.
The search has 2989 significant, unique peptide sequences, of which 2860 are in Unipept. The application essentially replaces conventional protein inference with LCA mapping. This produces summary graphs like the below treeview, where the thickness of the line indicates the amount of unique peptide evidence at each taxonomic level. The Chordata peptides are down to bovine trypsin and human proteins, while the bacteria are human gut microbes. There are lots of options for various hierarchical outlines, heatmaps and Gene Ontology annotations.
A lot of added value comes from simply exporting the results from Unipept as CSV. Sort by the ‘species’ column and the table lists all species uniquely identified by at least one significant peptide match, as shown in the screenshot below.
What about the 129 unmapped peptide sequences? These are peptides unique to the draft genomes and translated ORFs. As more genomes are sequenced and added to UniProtKB, the number will reduce. But a few peptide matches are actually non-tryptic. HTTQDHTTTPVLDHVNAAATAN is a C-terminal peptide from a translated ORF and gets a strong match. Mascot also finds a match in a hypothetical Bacteroides uniformis protein to the longer tryptic peptide that does exist in Unipept, HTTQDHTTTPVLDHVNAAATANSISTDYNEAYFHVSPSVGVR. If the sample contains many non-tryptic or semi-specific peptides, it’s something to keep in mind.
Family-level analysis
Unipept maps peptides directly to taxa and GO terms, and it’s not straightforward to get a listing of, say, all rubredoxin hits. You could drill down to a protein list by looking at individual peptide sequences in Unipept, but it’s easier to reanalyse just the peptides in the protein family. From the CSV file, select peptide sequences where prot_hit is 19, save in a text file and create a new assay in Unipept. The screenshot below illustrates the GO terms associated with protein family 19.
Unipept has a scriptable API, so it wouldn’t be hard to automate family-level analysis from the Mascot CSV export.
There’s currently no mechanism in Unipept to limit the range of species used in the analysis. For example, EGYPEVGLYYEK is unique to rubredoxin Dorea longicatena in the Mascot search results. But pasting the sequence in Unipept’s tryptic peptide analysis displays a list of 135 proteins. Many of these are from species not usually found in the human gut, which is why they were deliberately excluded from the Mascot search. Maybe a future version will allow the database lookup to be restricted to a list of taxonomy IDs?
Keywords: export, metaproteomics, protein inference