|
To view this email as a web page, click here. |
 |
|
Welcome
A new standardized method can unravel counterfeit cashmere.
This month's highlighted publication demonstrates some of the benefits and risks of protein carriers.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
We are pleased to announce a that the release candidate for Mascot Server 2.8 is now running on our public web site.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Counterfeit cashmere
Adulteration of cashmere products has become a worldwide problem, with cheaper substitutes such as sheep, yak and even rat being used in counterfeit sweaters and scarfs. The current approved methods have limitations such as hard to detect chemical treatments (microscopy) or have reproducibility problems (reductive alkylation LC/MS).
As an efficient alternative, a new ISO international standard method has been developed based based on research by the Biological Resource Center of the National Institute of Technology and Evaluation in Japan. The fiber samples are pulverized, then digested directly with trypsin with no reductive alkylation. One marker peptide per species is selected based on reproducibility and clear LC separation. The new sample preparation produces consistent and reproducible peptide intensities, unaffected by dyes or physical complexity.
A Unipept search shows the cashmere and sheep wool peptides to be unique to those species, while the yak peptide is shared with cows. Besides being able to identify cashmere, sheep, yak, camel, alpaca, and angora rabbit, the method also provides for the determination of blending ratios.
Go here for details about how to authenticate your sweater.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
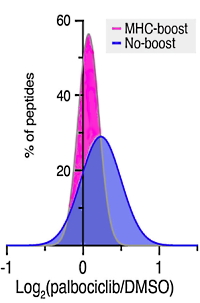
Quantitative consequences of protein carriers in immunopeptidomics and tyrosine phosphorylation MS2 analyses
Lauren E. Stopfer, Jason E. Conage-Pough, Forest M. White
Molecular & Cellular Proteomics Published online May 27, 2021
The authors have carefully examined the protein carrier approach to enhancing low level peptide signals, looking at peptide MHC profiling as well as tyrosine phosphorylation. Using 6-plex and 10-plex TMT labelling and MS2 quantitation, they assessed the number of unique peptide ID's as well as the intensities and fold changes for samples with and without carriers.
Including a protein carrier ("boost") increased the number of pMHC IDs: from a single injection, 3,176 unique pMHCs were identified in the boost sample, whereas 1,619 were identified in the no-boost. However, investigating proteins in pathways known to be perturbed by CDK4/6 inhibition, they found that the use of the protein carrier altered the quantitation. No pathways, including the three enhanced in the no-boost analysis, showed significant enrichment using the pMHC-boost data.
Similarly, looking at pTyr using Pervanadate to halt tyrosine phosphatase activity and increase tyrosine phosphorylation, they found that boost analysis identified more unique pTyr peptides compared to the no-boost analysis (3971 vs. 556). The boost analysis, however, contained just 163 pTyr peptides quantifiable across all samples versus 327 in the no-boost analysis, reducing overall data quantity. The authors urge careful assessment before using protein carriers in quantitation.
|
 |
 |
 |
 |
|
Mascot Server 2.8
The release candidate for Mascot Server 2.8 is now running on our public website. Please give it a try. Improvements include:
- New statistical model for error tolerant searches. Mascot assigns expect values to ET matches and (if the search is auto-decoy) estimates FDR.
- Increased Percolator sensitivity. Mascot ships with a new Percolator feature set, and the Percolator version has been updated.
- MS/MS searches are faster. Singly threaded file processing steps have been multithreaded, yielding 20-35% speed improvement.
Once testing is complete, customers under warranty or a support contract will receive the new version for free. If you have an earlier version, updating to version 2.7 comes with one year of Premium Support, giving you an update to version 2.8 as soon as it is released at no additional cost. Simply e-mail support@matrixscience.com for a quote.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|