|
To view this email as a web page, click here. |
 |
|
Welcome
Improvements to Mascot Distiller provide faster, more accurate label-free quantitation.
This month's highlighted publication demonstrates the rapid labeling and analysis of dynamic protein complexes.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month concerns using retention time as a Percolator feature.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Faster label-free quantitation with Mascot Distiller 2.8
Improvements in label-free quantitation (using the relative intensities of extracted ion chromatograms), mean that results are returned 2 to 3 times faster with Mascot Distiller 2.8 vs. Distiller 2.7.1, and while using about a third less memory.
In Mascot Distiller 2.8, global time alignment for a consensus dataset is achieved by roughly aligning the separate files using their Total Ion Chromatograms (TICs) and a recursive cross correlation method adapted from signal processing. Calculated retention times can then be used as the starting point for XIC detection in individual raw files. This provides some useful advantages:
- Sample files are processed and searched individually, reducing the memory footprint. XIC detection is also carried out separately, which allows for more efficient parallelization.
- You no longer need to estimate an upper limit on the shifts in retention times between files.
To assess this new capability, a data set of 12 samples containing E. coli spiked into HeLa cells at 4 different levels was analyzed. A total of 2072 human and 317 E. coli proteins were identified at a 1% PSM FDR. For expected ratios of 5, 2 and 1.5, Distiller 2.8 yielded values of 5.54, 2.00, and 1.64, which compares well with results from other software.
Go here to read more details about how Distiller 2.8 can speed up your workflows.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
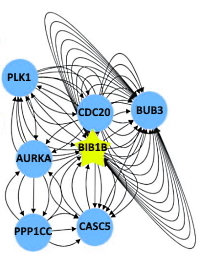
Mapping Proximity Associations of Core Spindle Assembly Checkpoint Proteins
Yenni A. Garcia, Erick F. Velasquez, Lucy W. Gao, Ankur A. Gholkar, Kevin M. Clutario, Keith Cheung, Taylor Williams-Hamilton, Julian P. Whitelegge, and Jorge Z. Torres
Journal of Proteome Research Published online June 4, 2021
The spindle assembly checkpoint (SAC) is essential for ensuring the fidelity of chromosome segregation during cell division. Here the authors have attempted to illuminate the regulatory factors involved in the SAC pathway, as misregulation can lead to tumorigenesis and resistance to chemotherapeutic drugs. Due to the dynamic nature of the associations between core SAC proteins and the complexes and subcomplexes that they form, it has been difficult to generate a view of their protein network.
The authors developed vectors for BioID2-tagged protein cell lines, where the tag is fused to each of the five SAC core proteins. When expressed in cells these proteins are then induced to biotinylate interacting and proximate proteins. The biotinylated proteins are purified on streptavidin beads, on-bead digested with trypsin, and analyzed by 2D LC-MS/MS. Relative quantitation was accomplished using protein coverage and the emPAI protocol.
For each of the core SAC proteins, a proximity protein association map was generated using custom R scripts. These proximity maps were then integrated to create a view of the core SAC network. Coupling the proximity maps/network with curated functional databases provided a system-wide analysis of the associations.
Though the analysis reiterated many of the core SAC protein−protein interactions, subcomplexes, and complexes that had been previously described, it also identified many novel associations.
|
 |
 |
 |
 |
|
Mascot tip
If retention time, using the RTINSECONDS parameter, is included in the peak list submitted to Mascot Server, it can be used as a feature by Percolator, which compares experimental and calculated RT values and includes this in the scoring.
This feature is disabled on installation but can be enabled by setting PercolatorUseRT to 1 in the Options section of mascot.dat. However, retention time calculation in Percolator is very time consuming and the sensitivity improvement is only marginal in most cases, so we advise against turning it on as a global default. If Percolation seems to take a very long time, you might want to check this setting.
Better to investigate the benefit of using the retention time feature for individual search results by manually adding the argument percolate_rt=1 to the report URL.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|