Error tolerant searches now show statistical significance
The latest release of Mascot Server introduces some important changes to error tolerant searches. Matches from the second pass search now have expect values attached, indicating confidence levels. These are either estimates based on counting trials or empirical values derived from searching a decoy database.
If you are not familiar with the error tolerant search, now is the time to give it a try. It is the most efficient way to find unsuspected modifications, non-specific cleavage products, and SNPs. Simply check the error tolerant option on the search form. A standard, first pass search is performed using the search parameters specified in the form. From the results of the first pass search, all of the database entries that contain one or more significant peptide matches are selected for the error tolerant, second pass search. These proteins are searched with relaxed enzyme specificity, while iterating through a comprehensive list of chemical and post-translational modifications, together with a residue substitution matrix. At the completion of the second pass search, a single report is generated, combining the results from both passes.
In Mascot Server 2.8, we have added two new search parameters. First, whether to search a decoy database or not. Without a decoy, significance thresholds and expect values are derived from counting trials – the number of candidate peptides that are tested for a match. This estimate is not always accurate; particularly when there is something wrong with the database or search parameters, making a large fraction of potential matches unavailable. Ticking the checkbox to search a decoy database may double the search time, but it gives a solid, empirical basis for the statistics.
The other new choice is the required false discovery rate (FDR) for the complete set of results. The reason this is asked for up front is that the required FDR determines the set of proteins selected for the second pass search. For example, the first pass search might identify significant peptide matches to 500 proteins at an FDR of 5%, and these are sent through to the second pass. If the FDR was reduced to 1%, the number of proteins selected for the second pass might drop to 400. Although the FDR can be tweaked at the report stage, this will not give perfectly identical results to setting the required FDR in the search parameters. (If the required FDR is set to no target, the default significance threshold is used to determine which peptide matches are significant and hence which proteins go through to the second pass, as in previous versions of Mascot Server.)
The target and decoy proteins are treated as pairs. After the first pass search, when proteins are selected, each significant match, whether target or decoy, causes the relevant pair of target and decoy proteins to be selected for the second pass. This means that the target and decoy databases are of identical size and contains all significant peptide matches (PSMs) from the first pass. The required FDR is applied independently to the results from the first and second pass searches. Since this is based on counts of PSMs, if it can be achieved for the results from each pass, then it will also be true for the combined results.
If a query gets a significant match in the first pass search, this is what we report, and we blindly discard the second pass results for this query. Sometimes, this means a stronger match is missed, but to do otherwise would be statistically dishonest. For example, if the significance threshold for a particular query in the first pass search corresponds to a score of 40, and we get a match with a score of 52, this is what we report, even if the second pass search would give us a match with a score of 80. This is not ideal, but the alternative is to burden all matches with statistics based on both passes. To illustrate why this is a problem, imagine we look at the second pass results and find nothing better. Now, we have a larger search space and the score threshold has increased to 55, so we have to discard our first pass match with a score of 52 because it is no longer significant. Sometimes you win, sometimes you lose.
As a real-life example, we have taken LTQ-Orbitrap Velos data acquired by the Medical University of Graz and deposited in PRIDE project PXD002726. Full details can be found in an earlier blog article. The search results are here.
Search parameters can be inspected by expanding the relevant section in the report header. It is also important to consider the composition of the local Unimod database. In this case, it was the latest update, but modifications classified as isotopic labels and those with deltas greater than 1000 Da were excluded from the error tolerant search. This reduced the number of modifications from 1499 to 1045.
Expand the Sensitivity and FDR section to see that the sensitivity at 1% FDR for PSMs was 4279. The protein FDR doesn’t look very pretty, but this is a consequence of the 42 decoy PSMs being scattered randomly across 20 decoy proteins. If we increase the Min. number of sig. unique sequences from 1 to 2 and choose Format, the protein FDR drops to a more satisfactory 0%.
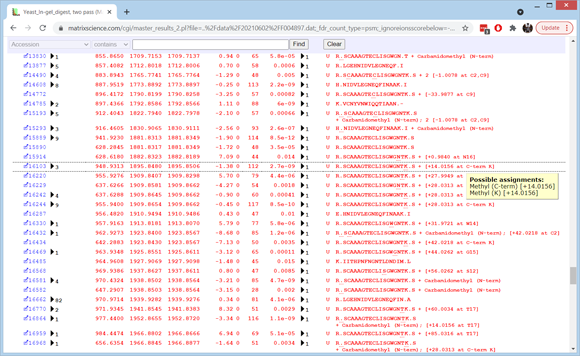
If you want to see how many additional matches were obtained in the second pass search, make a note of the PSM count (4279) then set Error tolerant matches to None and choose Format. The count drops to 2972, so there were 1307 new matches at 1% FDR. Set Error tolerant matches back to Reliable and choose Format again. Expand the Modification statistics section of the header to get an overview of where the error tolerant matches come from. Non-specific cleavage is up near the top of the list, along with deamidated, ethyl, methyl, and guanidinyl. Most of the deamidation and guanidination will be artefactual. Much or all of the ethyl is really dimethyl, which is unusually abundant because trypsin is methylated to reduce autolysis.
To see details of individual matches for the top 10 protein families, choose Expand all at the bottom of the page. The first family are human keratin contaminants. Scroll down to the matches to R.TSQNSELNNMQDLVEDYK.K. There is a first pass match to the oxidised peptide for query 18970 and a second pass match to the oxidised and deamidated peptide for query 18999. Both have a score of 99 but the first pass match has an expect value that is lower by a factor of 300, reflecting the smaller search space. If you hold the cursor over the value in the rank column, you can see that the score threshold is 18 for the first pass and 39 for the combined passes.
Although this example is dominated by artefactual modifications, and it is hard to spot any post-translational modifications, it serves to demonstrate how the new features work to provide confidence in the matches and modifications being reported. For more detailed information, refer to the help page for error tolerant searches.
Keywords: delta mass, error tolerant, FDR, modification, statistics