|
To view this email as a web page, click here. |
 |
|
Welcome
If you are planning to attend ASMS in Philadelphia, we'd be delighted to meet you at booth 327 in the exhibition hall. Since international travel is still difficult, our user meeting presentations will be online only. More details in our next newsletter.
Enhancements in error tolerant searching provide improved confidence through robust statistics.
This month's highlighted publication shows a streamlined approach to finding antimicrobial peptides in hot peppers.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to export ion mobility values to peak lists when using Mascot Daemon.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Enhanced confidence for error tolerant searching
Error tolerant searching is the most efficient way to find unsuspected modifications, non-specific cleavage products, and SNPs. In Mascot Server 2.8, we have implemented improvements to error tolerant searches where matches from the second pass search now have expect values attached, indicating confidence levels.
When a decoy search is specified, the process operates like this:
- A standard, first pass search is performed of target and decoy databases
- Target and decoy proteins are treated as pairs. After the first pass search, each significant match, whether target or decoy, causes the relevant pair of target and decoy proteins to be selected for the second pass
- The new target and decoy databases are searched with relaxed enzyme specificity, while iterating through a list of PTMs and residue substitutions
- After the second pass search, a report is generated, combining the results from both passes.
The required false discovery rate (FDR) is specified up front because this determines the set of proteins selected for the second pass search.
If a query gets a significant match in the first pass search, this is what we report, and we blindly discard the second pass results for this query.
Go here to learn more about these capabilities and how the various parameters affect the search results.
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Too Hot to Handle: Antibacterial Peptides Identified in Ghost Pepper
Kevin D. Culver, Jessie L. Allen, Lindsey N. Shaw, and Leslie M. Hicks
J. Nat. Prod. 2021, 84, 2200-2208
Though hot peppers are known primarily for their capsaicinoids and the resulting heat, they do host a number of other bioactive compounds that the authors have investigated. They began by mining genomic data for antimicrobial peptides (AMPs).
They used SignalP to predict and cleave signal peptides from the translated proteome in silico, and then analyzed the remaining peptides with Cysmotif Searcher, which identified any cysteine motifs. This provided 115 sequences identified as potential AMPs. The ghost pepper extract was fractionated and analyzed by LC-MS/MS to identify 15 previously uncharacterized AMPs from this list.
Fractions were assayed against a number of pathogens and high-responding fractions were identified. The authors then applied their PepSAVI-MS statistical analysis package to filter compounds to include only those likely contributing to the observed bioactivity. These signals were further investigated by de novo sequencing to elucidate the structure of two novel AMPs which showed low micromolar IC50 antimicrobial activity against several gram-negative bacteria.
|
 |
 |
 |
 |
|
Mascot Tip
Mascot Distiller 2.8 includes support for Bruker timsTOF data. If you are using Mascot Daemon to batch process raw files, and wish to include precursor ion mobility values in the peak lists, you need to enable this using the following procedure:
- Log into Windows as a user with Administrator privileges
- Type cmd into the Windows search bar
- Right click Command Prompt or cmd.exe and choose Run as Administrator
- Navigate to the directory containing MascotDistiller.exe (by default this will be C:\Program Files\Matrix Science\Mascot Distiller)
- Run the following command (all on one line):
MascotDistiller.exe -batch
-daemonMGFIonMobilityParam 1
During quantitation, ion mobility filtering is automatically enabled for any search results with the precursor ion mobility values exported in the peak lists. More information can be found in the Mascot Distiller help and in this blog article.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
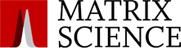 |
 |
|
|