|
To view this email as a web page, click here. |
 |
|
We wish you a Happy New Year.
Hardware for Mascot Distiller can have a large effect on performance, and we have some guidance on this.
This month's highlighted publication describes a new serological assay for determining SARS-CoV-2 antibodies.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month explains how to make the Protein Family Summary the default report for all MS/MS searches.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Choosing hardware for Mascot Distiller
A common question with Mascot Distiller regards the specifications of the CPU, RAM, and disk drives for the workstation on which it will be installed. We have some suggestions on how to choose.
Many processing steps in Mascot Distiller are multithreaded, speeding up complex tasks by parallelizing the work. Unlike Mascot Server, Mascot Distiller is not licensed by the core and can use all the CPU threads available to it to parallelize these tasks.
We ran a series of tests increasing the number of threads available to Mascot Distiller from 1 to 12 for peak picking, quantitation and de novo sequencing. The speed increased by a factor of 6-8x for each of these applications as the number of threads increased. The number of CPU cores is not the only factor which will determine overall processing time as the processes can hit other limiting factors. But as a general rule, for many people a modern 8-core CPU will give good overall performance for the cost.
As a rule of thumb for RAM, we'd recommend having at least 16GB, but 32GB or more if you're processing very large datasets.
Regarding hard drives, in the benchmarked peak picking, quantitation and de novo searching tests, we typically see a 5% improvement in performance using solid state drives over magnetic.
Go here to read more about hardware for Mascot Distiller.
|

|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
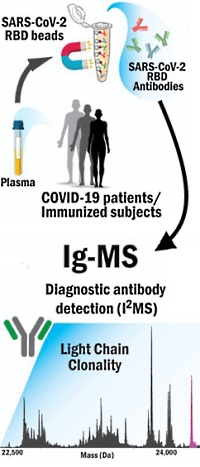
Next-Generation Serology by Mass Spectrometry: Readout of the SARS-CoV-2 Antibody Repertoire
Rafael D. Melani, Benjamin J. Des Soye, Jared O. Kafader, Eleonora Forte, Michael Hollas, Voislav Blagojevic, Fernanda Negrao, John P. McGee, Bryon Drown, Cameron Lloyd-Jones, Henrique S. Seckler, Jeannie M. Camarillo, Philip D. Compton, Richard D. LeDuc, Bryan Early, Ryan T. Fellers, Byoung-Kyu Cho, Basil Baby Mattamana, Young Ah Goo, Paul M. Thomas, Michelle K. Ash, Pavan P. Bhimalli, Lena Al-Harthi, Beverly E. Sha, Jeffrey R. Schneider, and Neil L. Kelleher
J. Proteome Res., published online: December 08, 2021
The authors sought to improve serological testing by developing a more precise assessment of immune status relative to SARS-CoV-2. Their new method, Ig-MS, isolates antibodies against a specific target from the plasma of patients and breaks them down into their heavy and light chains (HC and LC). The HC and LC fragments are then analyzed by individual ion mass spectrometry (I2MS), an approach that generates mass distributions for heterogeneous protein samples using ˜500 fold more dilute samples than classical protein MS analysis.
They created bait for binding and enriching antibodies using magnetic beads covalently attached to the SARS-CoV-2 spike protein receptor-binding domain (RBD), and verified the assay performance with COVID-19 convalescent plasma, showing >90% recovery.
Following antibody enrichment, two distinct workflows were utilized. In workflow 1, Ig-RBDs were eluted intact, combined with the standard mAb, and fully reduced/denatured to liberate HC and LC proteoforms. In workflow 2, Ig-RBD were digested with IdeS protease while still attached to the beads, generating F(ab′)2 and Fc. The protease and Fc species (containing HC glycosylation) are then washed away. Eluted F(ab′) 2 were denatured and reduced to yield the LC and the Fd domain (˜25-28 kDa).
They created two new metrics from these methods - Ion Titer (IT) is similar to an ELISA titer and uses the intensity of all LC peaks relative to the mAb standard. The second is the Degree of Clonality (DoC), a measure of the complexity of all LC clones (proteoforms) in the mixture, with higher numbers reflecting a more complex Ig-MS spectrum.
Though correlation of DoC with standard ELISA and bioluminescence was low, they did find significant positive correlation of IT obtained with workflow 2 and the ELISA titers.
|
 |
 |
 |
 |
|
Mascot Tip
The Protein Family Summary was new in Mascot Server 2.3, almost 12 years ago. It displays MS/MS search results a page at a time, allowing the results of even the largest searches to be browsed without client-side memory or browser limitations. Proteins are grouped into families using hierarchical clustering, making protein inference more intuitive and more accurate. To avoid upsetting established workflows, the new report was only the default for searches of more than 300 MS/MS spectra, and the earlier Peptide Summary report continued to be the default for smaller searches.
Since then, many new features have been implemented in the Protein Family Summary, and the Peptide Summary cannot be used for anything other than 'vanilla' searches. In Mascot 2.8, we decided the time had come to make the Protein Family Summary the default for all MS/MS searches.
If you are using an earlier version, and would like to follow suit, it is a simple configuration change. Using the Configuration Options module of the Configuration Editor, change the value of ProteinFamilySwitch from 300 to 1 and choose Apply. If you ever need to see search results in the earlier format, follow the 'Try the peptide summary' link in the report header.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|