|
To view this email as a web page, click here. |
 |
|
Welcome
We enjoyed seeing many of you at the ASMS meeting in Minneapolis last week. If you missed the presentations in our user meeting describing the latest developments in Mascot Server and Distiller, slides and speaker notes can be found below.
This month's highlighted publication shows the use of long-read RNA-seq to construct protein databases that can better identify splice isoforms.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month discusses an aspect of False Discovery Rate
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Identification of Protein Isoforms Using Reference Databases Built from Long and Short Read RNA-Sequencing
Aidan P. Tay, Joshua J. Hamey, Gabriella E. Martyn, Laurence O. W. Wilson, and Marc R. Wilkins
J. Proteome Res., published online May 25, 2022
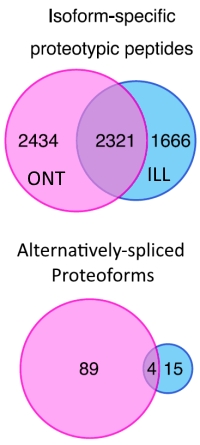
Alternative splicing leads to transcripts that vary in their coding and untranslated regions, resulting in distinct protein isoforms with potentially different functions. The number of functionally important protein isoforms arising from alternative splicing remains mostly unknown, so the authors have investigated the use of long- vs short-read RNA-sequencing data to build custom protein databases. Since short reads can map to more than one genomic location and will rarely span across several splice junctions, they posited that the use of long-read RNA-seq should improve the identification of protein isoforms.
They used a proteomic data set for human K562 cells that they obtained from the ProteomeXchange, and searched this against protein databases constructed from RNA-seq data derived from Illumina-based short-read or the Oxford Nanopore long-read RNA-sequencing platforms.
The results showed a good deal of complementarity, but searching with the long-read database identified 4755 isoform-specific proteotypic peptides while the short read database identified 3987. They also searched for protein isoforms arising from alternative splicing and found 93 protein isoforms from 39 genes with the long-read database and 19 protein isoforms from 10 genes with the short-read database.
|
 |
 |
 |
 |
|
Mascot Tip
According to Wikipedia, the average population density of the United States is around 36 people per sq km. Even someone with a very shaky grasp of mathematics would understand that this figure does not hold true for selected regions. The density within large cities is orders of magnitude higher than the average while that of Alaska or Death Valley is much lower.
It's a similar case with False Discovery Rate, but for some reason this is less intuitive. The FDR for a complete search result is an average value. We can use target-decoy to adjust the global FDR to precisely 1%, but this will not be the FDR for a sub-set of matches, such as long peptides or short peptides or peptides with variable modifications.
A series of blog articles discusses this and related issues and gives some numbers. For the typical search used as an example, at an FDR of 1%, only 3% of target matches contained any variable modifications compared with 41% of decoy matches. If you want to report an FDR for a sub-set, such as modified peptides or non-specific peptides, it has to be based on counts of this class of matches, not all matches.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|