|
To view this email as a web page, click here. |
 |
|
Welcome
A crosslinking analysis yields many unexpected disulfide bonds in the SARS-CoV-2 spike protein.
This month's highlighted publication describes an approach to use N-terminal peptides to elucidate protein products from non-coding regions.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month addresses disaster recovery: what to back up and how to restore Mascot Server, Distiller and Daemon.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
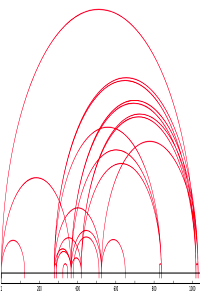
28 Disulfide bonds in the SARS-CoV-2 Spike protein?
With over 3000 papers on the proteomics of the SARS-CoV-2 virus, much is known, especially about the immunologically important spike protein that is involved in the initial binding to the target cells. A recent spike protein study investigated the protein sequence, glycosylations and disulfide bonds and then used the information for 3D modeling and interaction studies, aiming ultimately to facilitate immunogen design.
We used the crosslinking feature of Mascot Server to reanalyse the data set available on PRIDE.
The purified mimetic protein was not reduced but was alkylated, then submitted to either multi-enzymatic digestion with trypsin, Lys-C and Glu-C, or digested with α-Lytic Protease. This was followed by LC/MS/MS using ETD. Since crosslinked peptides tend to have higher precursor masses and charge states than typical peptides, we used Mascot Distiller to deconvolute and decharge the fragment ions, producing singly charged MH+ values.
Searches of both enzymatic preparations identified far more disulfide bonds than expected from reading the paper. Mascot Server identified 28 disulfide bonds of which 8 were previously recorded in UniProt, while the authors only report a single disulfide bond between Cys15 with Cys136. Since this was a mimetic immunogen rather than material extracted from a SARS-CoV-2 virus preparation, it is not clear if the 20 additional disulfide bonds can occur naturally. Either way, it will be interesting to see if additional disulfide linkages are reported in future investigations.
Read more about the spike protein crosslinks in our blog.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
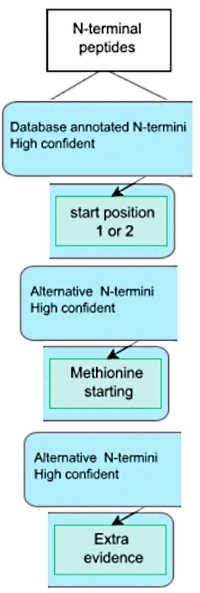
Limited evidence for protein products of non-coding transcripts in the HEK293T cellular cytosol
Annelies Bogaert, Daria Fijalkowska, An Staes, Tessa Van de Steene, Hans Demol, Kris Gevaert
Molecular & Cellular Proteomics, July 1, 2022, in press
The authors investigated the discrepancy between the number of protein products from non-coding regions predicted from RNA analyses and the number of unambiguously detected (non-coding region) proteins typically reported from MS-based proteomics.
They examined the cytosolic extracts from human embryotic kidney cells (HEK293T) and enriched the N-terminal peptides. In theory, most proteins can be identified by their N-terminus alone and the N-termini are ideal proxies for studying protein variants. First, the primary amines in the cytosolic proteins were acetylated with a heavy acetyl group to distinguish in vivo N-terminally acetylated from in vivo free N-termini. Three proteases – trypsin, chymotrypsin, and GluC – were used in parallel to increase coverage, and N-terminal peptides were concentrated by two subsequent HPLC fractionations with a TNBS reaction in between.
The LC-MS/MS data were searched against a custom database consisting of UniProtKB-SwissProt entries and UniProt isoforms appended with a Ribo-seq based protein database. Combining the data from the different proteases led to the identification of 2,896 distinct N-termini and 2,420 distinct proteins. The authors then applied a rule-based selection strategy that first removed internal and C-terminal peptides, followed by stringent selection based on N-terminal co-translational acetylation rules, the presence of an initiator methionine (and its processing), and evidence of translation by Ribo-seq. This left them with 19 potential novel proteins.
|
 |
 |
 |
 |
|
Disaster recovery
There are a number of potential issues that can cause your Mascot Server or other computer infrastructure to stop working. With disk or storage failure, replacing the device will allow you to reinstall Mascot Server and get back up and running quickly. Or if it is a "simple" failure like the system board, the hard disks can be moved to another computer. On a computer that suffers RAID controller failure, you will need identical or at least compatible hardware in order to access the data again. For older systems this can be difficult or expensive to obtain.
In the worst case, you may need to do a full recovery from backups. This can be an involved procedure, but it is straightforward as long as you have backed up key configuration and data files. These are described in a new help page Disaster recovery on our website.
In short, back up your raw data, Distiller project files (rov files), Mascot Server configuration files and database search results, and the Daemon parameter files and task database. Restoring from backup can be done by reinstalling the software and copying backed up files to the right places.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|