|
To view this email as a web page, click here. |
 |
|
Welcome
Fractionated label-free quantitation is now supported in Mascot Distiller.
This month's highlighted publication describes how peptide biomarkers are used to identify flounder, plaice and sole.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Certain Linux systems set a low limit on the maximum number of open files, which can cause odd behaviour. Mascot tip of the month has a solution.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
Fractionated label-free quantitation (LFQ) with Mascot Distiller
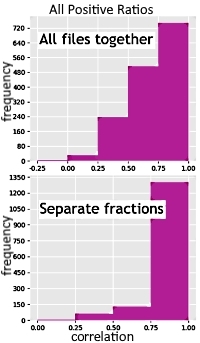
Label-free quantitation of fractionated samples is supported by Mascot Distiller since version 2.8.2, released earlier this year. When Distiller aligns raw files in the time domain, it creates a consensus and aligns each raw file to it. The alignment is used as a starting point for peptide XIC detection for any peptide match that is present in one file and absent in another. With fractionated samples, Distiller now creates multiple consensuses – one for each fraction. Files are aligned to the consensus for their assigned fraction, instead of trying to align everything to a single consensus. This reduces noise and makes quantitation faster, as the system doesn't waste time looking for a peptide in irrelevant fractions.
We reanalysed a GelC-MS data set with samples from HeLa (wild type) and HeLa KO30 cells to demonstrate the effect. Protein extracts from each sample are run on a 1D gel, and the lanes are cut into slices, which are digested and run via LC-MS/MS. Quantitation was run on the dataset either aligning all files to a single consensus, or just aligning matching slices (fractions). In both cases, 1417 peptides were quantified. However, only 569 of those pass the quality thresholds if fractionation is ignored, while 1137 peptides are accepted when fractions are aligned correctly. Additionally, time alignment with fractions enabled took only 11 minutes to complete compared with 26 minutes if fractionation is ignored.
Our blog has a more in-depth description of fractionated LFQ.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Peptide mass fingerprinting of preserved collagen in archaeological fish bones for the identification of flatfish in European waters
Katrien Dierickx, Samantha Presslee, Richard Hagan, Tarek Oueslati, Jennifer Harland, Jessica Hendy, David Orton, Michelle Alexander and Virginia L. Harvey
R. Soc. Open Sci., 2022, 9, 220149
The authors investigated flatfish bones from 3 archaeological sites around the North Sea area to better understand shifts in the environment, economy, fisheries, human diet, and social status throughout history. Since these species are difficult to identify by conventional morphological analyses, many questions remain unanswered about their exploitation and how it might have changed over time.
They employed ZooMS (Zooarchaeology by Mass Spectrometry) which uses peptide mass fingerprinting (PMF) to identify collagen 'Type I' preserved in bone tissue, as well as LC-MS/MS to assign sets of biomarkers for taxonomic identification. Using modern flatfish bones from museum and fresh specimens caught in the North Sea and surrounding areas, the collagen was extracted from the bones, analyzed by MALDI-TOF and LC-MS/MS and searched against a local database with 151 published fish collagen sequences obtained from NCBI Blast.
Eight collagen peptide markers suffice to identify at least 18 different species from the 202 archaeological flatfish bones from the 3 sites. In addition to seeing the transition from freshwater and estuarine exploitation to marine fishing (from the 7th to 16th centuries), this methodology could be used to address the misidentifications in the food chain of wild-caught fish.
|
 |
 |
 |
 |
|
System limits on Linux
We've recently had reports of odd Mascot behaviour on certain Linux distributions. The main symptoms are that Mascot is suddenly not able to bring a new or updated database online, and the monitor log, error log or search log stop updating. Increasing Mascot's soft limit MaxDatabases makes no difference, and file ownership and permissions don't explain it. Restarting the Mascot monitor service (ms-monitor.exe) or the PC sometimes helps, sometimes not.
The issue is caused by a system limit on the number of open files. The Mascot monitor service keeps open six files for each active database, plus it needs to open and close the log files and certain control files as needed. When a database is being updated, compressing the new version requires up to 10 more files to be open. As soon as the open file limit is reached, the operating system will prevent the monitor service from opening any new files, including log files.
You can find the current limit by inspecting the /proc/[PID]/limits file, where [PID] is the process ID of ms-monitor.exe. If "Max open files" is 1024 or less, we recommend increasing it. The correct way depends on how you start ms-monitor.exe. If you launch ms-monitor.exe on the command line, then the limit is set in the current user's .bashrc or .profile by running ulimit before starting ms-monitor.exe. If you launch ms-monitor.exe as a system service, then the limit nofile (number of files) is typically set in /etc/security/limits.conf. On some systems, it may be set with DefaultLimitNOFILE= in /etc/systemd/system.conf. After changing the limit, restart ms-monitor.exe.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|