|
To view this email as a web page, click here. |
 |
|
Welcome
Mascot Distiller provides many built-in and customisable quantitation reports.
This month's highlighted publication examines the diverse array of PTMs on tubulin and their effect on function.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Tip of the month concerns the TMTpro quantitation method shipped with Mascot.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
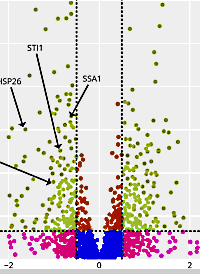
ANOVA, Box-Plot, clustering reports, volcano plots and more
Mascot Distiller ships with many graphical and tabular quantitation reports. In the latest version, reports are written in Python and use Mascot Parser to access the search and quantitation results. This additional flexibility has allowed us to add options to generate ANOVA, Box-Plot, various clustering reports and volcano plots, and a new option to export protein and peptide quantitation data for use in tools such as R and Perseus.
Reports are run using a wizard interface, where you specify the format and parameters. You can export static images or use the Javascript-enabled interactive report, which allows zooming, filtering, annotating and producing publication-ready graphics. You can also add custom reports to the system by writing your own Python code.
To demonstrate the new reports, we used a label-free quantitation dataset that looked at the role of alanyl-tRNA synthetases in S. cerevisiae. We reprocessed the raw data using Mascot Distiller, searched the generated peaklists with Mascot Server using the search settings described in PRIDE, then carried out quantitation in Mascot Distiller. We created a variety of reports that help to reveal biologically relevant information from the data.
You can see the example plots and reports in our blog.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Combinatorial and antagonistic effects of tubulin glutamylation and glycylation on katanin microtubule severing
Ewa Szczesna, Elena A. Zehr, Steven W. Cummings, Agnieszka Szyk, Kishore K. Mahalingan, Yan Li, Antonina Roll-Mecak
Developmental Cell, 57, 2497–2513, November 7, 2022
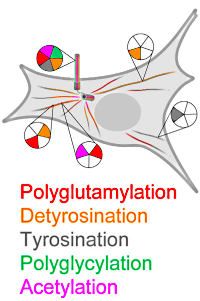
Microtubules – polymers of α,β-tubulin heterodimers – give structure to cells, build morphologically varied structures such as spindles, cilia, and flagella, and serve as tracks for intracellular transport. Microtubules have chemically diverse posttranslational modifications, and these PTMs act as part of a "tubulin code" that helps regulate the recruitment and activity of microtubule effectors.
The authors investigated how the PTMs concentrated on tubulin tails regulate katanin, a tubulin effector that extracts tubulin subunits from microtubules and severs it along its length. They utilized unmodified human tubulin and employed recombinant glutamylases to modify it in vitro.
At a mean glutamate number of ~3 on α-tubulin, severing is enhanced ~5-fold, and at a level of ~7, severing is enhanced ~9-fold compared with unmodified microtubules. Additionally, microtubule recruitment increases with increasing glutamylation of both α- and β-tubulin. MS/MS mapping of the glutamylation sites on the β-tail show preferential glutamylation at E441, followed by E438, and to a lesser extent at E439, E442, and E443.
The authors also examined whether α-tubulin detyrosination (removal of the terminal tyrosine) regulates katanin and showed that detyrosination decreases katanin microtubule binding while also decreasing severing. They looked at the combinatorial effect of these two modifications and found that polyglutamylation on either the α- or β-tail overcame the inhibitory effect of detyrosination. Glycylation decreases both katanin microtubule binding and severing with inhibition proportional to the glycine number, while acetylation does not affect katanin microtubule association or severing.
|
 |
 |
 |
 |
|
TMTpro quantitation methods
Mascot Server 2.8.1 added quantitation methods for TMTpro 16-plex and 18-plex. Unfortunately, the two TMTpro methods have an error. The 'required' attribute is checked for the fixed modification group, which means a peptide is only quantified if it has a lysine and the lysine has TMTpro fixed mod and the N-terminus has TMTpro fixed mod. If a peptide has no lysine, then according to the quantitation method, it's not quantifiable (it's missing a required fixed mod). Usually, the 'required' attribute should only be checked if the protein was labelled prior to digestion with trypsin, where roughly half the peptides would be unlabelled and should be excluded from quantitation. If labelling is done after digestion, 'required' should be unchecked.
The configuration error doesn't affect the database search itself, as Mascot applies fixed mods from the modification group the same way whether 'required' is on or off. At the end of the search, the search report calculates the quantitation ratios for each peptide matching the fixed modification group. The configuration error affects the report's decision whether a peptide is quantifiable.
Apologies we did not catch the error in beta testing. The default configuration will be fixed in the next version of Mascot, and the fix in your local configuration is very simple. Go to the Mascot configuration editor, Quantitation, and edit the TMTpro 16-plex method. In the Method tab, open the "TMT fixed" modification group. Uncheck the Required checkbox, then click OK and Save changes. Repeat for TMTpro 18-plex. Alternatively, you can edit quantitation.xml in a good text editor and change the required="true" attribute to required="false" for the two quantitation methods.
With this 'one weird little trick', you've multiplied the number of your quantified TMTpro peptides!
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|