|
To view this email as a web page, click here. |
 |
|
Happy New Year!
TMT/TMTpro labeling sometimes generates tall complementary reporter ions, which can interfere with peptide identification. They can either be removed before the database search or used for quantitation.
This month's highlighted publication investigates the molecular effects of growing rye grass on Mars.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
If your Mascot Server suddenly stops working, one of the most common reasons is that the disk is full. This month's tip discusses solutions.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
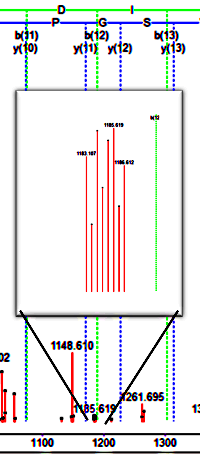
Complementary reporter ion clusters in TMT/TMTpro labeling
TMT/TMTpro labeling sometimes creates complementary ions in addition to or instead of reporter ions. Complementary ions are formed during fragmentation, where the precursor loses the reporter ion and carbon monoxide, leaving behind the peptide and the balance region of the label. If they are prominent in your data, their presence could interfere with peptide identification. This is because Mascot scoring penalises tall, unexplained MS/MS peaks. One solution is to remove the peaks before the database search. We wrote a script that removes the complementary ions given an MGF file, to see if there was a change in the peptide score. In an example data set, about 15% of peptide matches were affected, and the score of these increased by 3-4 on average after removing complementary TMTpro peaks.
Rather than removing complementary ions, we can use them for quantitation. In some cases, there can be interference in the standard reporter ion region from co-isolating peptides that then fragment along with the target peptide and distort the reporter ion measurements. We extended the script so that it can convert complementary ion masses into reporter ion masses and replace the original reporter ion peaks prior to searching and quantitation.
We evaluated the script using data from a yeast cell lysate in 6 channels at 0:1:5:10:10:5:1:0 concentration which was mixed with a human cell lysate. The ratios reported for the peptides matching to a yeast protein are close to the expected ratios of 5:1, 10:1 and 10:5.
You can read more details in
our blog,
which also has a download link for the script.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
A molecular study of Italian ryegrass grown on Martian regolith simulant
Roberto Berni, Céline C. Leclercq, Philippe Roux, Jean-Francois Hausman, Jenny Renaut, Gea Guerriero
Science of the Total Environment 854 (2023) 158774
For a sustained human presence on Moon or Mars, one of the most important challenges is to produce food locally as an alternative to expensive cargo shipments from Earth. Growing plants also produces oxygen and phytochemicals. In this study, the authors tried to mimic space farming by growing rye on a Mars soil simulant.
Italian ryegrass (Lolium multiforum) plants were grown on potting soil and Martian regolith (soil) simulant (MMS-1) a total of 14 days, with leaves sampled at 7 and 14 days. Some of the plants were also cut at 7 days to understand how they would react, and to have an estimation of the growth in a hypothetical scenario where ryegrass is mown to obtain organic matter for regolith amendment. Gene expression analysis together with shotgun proteomics were performed on leaves and roots. Additionally, light microscopy observations were carried out on roots and leaves after 14 days of growth.
The plants were able to grow on MMS-1, but compared to potting soil, the leaves showed extensive yellowing of the tissues starting from the external margins of the lamina towards the central vein. For shotgun proteomics, 50 mg of ground leaf and root tissues were taken for each sample, lysed and the extracted proteins digested before analysis by LC-MS/MS. The data were searched against a database of the Pooideae subfamily of grasses. The high-abundance proteins with the largest fold change (FC ≥ 30) between potting soil and Martian simulant roots are involved in primary metabolism, translation and cytoskeletal organization.
|
 |
 |
 |
 |
|
Free disk space
Mascot Server is, like the name says, a server application. It can be installed on a laptop or workstation and used interactively, but very often it's installed on a system in a different room or building, out of sight. If Mascot suddenly stops working, one of the most common reasons is lack of free disk space. This can manifest in a variety of odd errors, such as: all searches end permaturely (may appear as a "crash"), Mascot complains about truncated input or missing search parameters, unable to create session file (when trying to log in to Mascot Security). It can be easy to miss the early warning signs if you don't regularly log in via remote desktop or SSH to the server. We recommend setting up an automated monitoring system, or even just a scheduled task/cron job, that alerts you when free disk space drops below 10%.
The three largest consumers of disk space are sequence databases ('sequence' directory), results files for database searches ('data' directory) and cache files created for reports ('data/cache' directory). The cache directory contains temporary files that can be recreated when needed. If you quickly need to free up space, deleting the contents of data/cache will help. A better solution for the medium term is to regularly delete cache files that have not been accessed recently. Mascot ships with a tool, tidy_data.pl, for this purpose. The instructions for enabling the script are in chapter 7 of the Installation & Setup manual; search for "tidy_data".
How long to store search results in the 'data' directory is something to discuss with your Mascot users. If searches older than 12 months are rarely or never accessed, you could set up a scheduled task to archive or delete results files older than 12 months. If you want to keep them available just in case, then tidy_data.pl can compress old search results to save some disk space. The Mascot results report automatically decompresses the file when it's next viewed.
Finally, the 'sequence' directory may regularly grow in size if you have set up automated database updates in Database Manager, or if users regularly add new databases. The solution is to deactivate and delete the largest databases that are no longer needed. It's also worth checking the 'incoming' directory for each database, as they may contain unused temporary files from failed download attempts.
|

|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|