|
To view this email as a web page, click here. |
 |
|
Welcome
Analyzing ancient proteins is very challenging, and we discuss tips presented in a recent review.
This month’s highlighted publication searches for potential biomarkers of endometrial cancer.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Keeping Mascot Server up to date is easy and has several benefits.
Please have a read and feel free to contact us if you have any comments or questions.
|
|
|
|
 |
 |
 |
|
The challenges of ancient proteins
A recent review on paleoproteomics has highlighted a number of unique challenges that arise with the sample processing and data analysis of these ancient proteins.
The number one issue is sample collection and preparation, specifically being able to detect any protein at all. Anything that has been surrounded by unfavorable matter like soil, microbes or water for hundreds of years (or millions of years) may have very little protein left, and may be heavily modified due to diagenesis.
The next big challenge is with protein sequence databases that may contain few or no sequences for many historically or geographically important species, like local vegetables, mammals, birds or fish. The issue is similar to what one encounters with metaproteomics.
Although you can try de novo sequencing, a better approach is to make more comprehensive sequence databases from as wide a variety of species as feasible.
A few big projects like the Earth Biogenome Project are doing exactly that.
Even if the target species is not in the database, it helps to have something from the same genus or family.
Many diagenetic modifications like deamidation are so common that they can be used to authenticate ancient proteins. If an identified protein does not have a certain degree of expected damage, it could be a lab contaminant.
Our blog
has more tips and suggestions for improving your results when hunting for ancient proteins.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
A Label-Free Proteomic Approach for the Identification of Biomarkers in the Exosome of Endometrial Cancer Serum
Eduardo Sommella, Valeria Capaci, Michelangelo Aloisio, Emanuela Salviati, Pietro Campiglia, Giuseppe Molinario, Danilo Licastro, Giovanni Di Lorenzo, Federico Romano, Giuseppe Ricci, Lorenzo Monasta and Blendi Ura
Cancers 2022, 14, 6262
The authors searched for key mediators of Endometrial Cancer (EC) released in serum exosomes that could serve as potential biomarkers of cancer development and progression. The exosomes are microvesicles that originate in the multivesicular body and measure 30–100 nm in diameter.
To characterize the exosomal proteome of EC patients’ sera, they first isolated exosomes from the albumin-depleted sera of 12 EC patients and 12 controls. They performed label free quantitation on the LC-MS/MS data with Mascot Distiller using the Replicate Protocol, where the relative intensities of extracted ion chromatograms for precursors in multiple data sets are aligned using mass and elution time. Of the initial 421 protein groups identified, 31 proteins showed fold changes >1.5.
To build a robust model, the authors selected 8 proteins with fold changes >3.5. This yielded a predictive model with a sensitivity of 83% and specificity of 71%. Separating out only Stage 1 EC versus advanced cases, the refined model achieved a sensitivity of 100% with a specificity of 86%.
|
 |
 |
 |
 |
|
Staying up to date
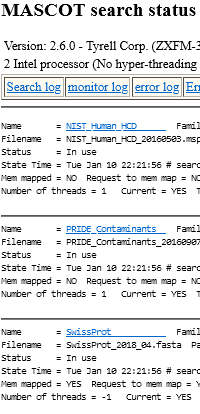
Are you running the newest version of Mascot Server that you have paid for? When a new version is released, and your licence is under warranty or support, you are entitled to a free update. We e-mail the new product key and download link to the Mascot contact we have on file. Sometimes these e-mails are overlooked or forgotten, or the person who received them is no longer responsible for the Mascot licence. Your installed version can be found easily: from your local Mascot home page, go to Database Status and the version is shown at the top of the page. If you are not sure about your support status or what is the newest version you could install, just e-mail your product key to support@matrixscience.com.
If you haven't updated in a while, there may have been good reasons to stick to the old version, for example being in the middle of a project. On the other hand, there are also good reasons to keep the software up to date. Staying on the latest version ensures you get bug fixes and updates when external circumstances change. Most recently, the UniProt REST API changed and NCBIprot reached critical size. The operating system itself may go out of support, and Mascot Server 2.6 and earlier are not likely to work on Windows 11/Server 2022. Every Mascot release has new features, but we don't change the software just for the sake of it. We try hard to maintain backwards compatibility and reproducibility, so it should always be safe to update.
Updating Mascot while staying on the same hardware is very easy. Just download and run the installer, which updates program files to the new versions. All configuration files, search results and sequence databases are kept. If the PC running Mascot is a few years old, moving to new hardware can easily double database search speed. Moving a Mascot licence is free of charge, and we can give you a 30-day temporary key to test the new system before decommisioning the old one. Detailed instructions are available for every scenario. If you'd like to switch platform (from Windows to Linux or vice versa), you can do so for free if your licence is under support. To get back under support, buy an update to the latest version, which comes with one year of Premium Support. It is always cheaper to update an existing licence than buy a new licence.
|
|
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
|
|
|
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|



|
 |
|
|