|
To view this email as a web page, click here. |
|
|
|
Welcome
Mascot Security can be set up so that colleagues can easily submit searches without the need to log in, while the admin side is protected with a password.
In this month's highlighted publication, the authors explored the relative performance of Mascot and PEAKS for identifying FPOP peptides and sites of modifications.
If you need software for your research lab, read on why Mascot gets over 2000 citations every year.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
Get a quote
|
 |
|
|
|
Mascot Security: easy searching with no login
Mascot Server includes a role-based access mechanism called Mascot Security.
This is often used to limit access to different parts of the system, for example requiring a password to change global settings that affect all Mascot Server users.
You can configure your Mascot Server so that the admin side is password protected but without having to log in to perform searches or view results. This could be useful for a research lab that is not sharing the results outside of the lab or otherwise has no need to restrict access to the search functions of the server.
To do this, log in as admin and go to the security editor. Enable the guest user, which is a user account that requires no password. Edit the guest permissions so that they can submit searches, view results, access databases and connect to the server with Mascot Distiller and Mascot Daemon. Since Security is enabled, only admins will be able to make changes in the configuration editors. Anyone else is "logged in" without a password as a guest user, who is prevented from accessing the global configuration.
Another way to get the same result is configuring Mascot to use web server authentication using Microsoft Active Directory or various single sign-on systems. When the user logs in to the operating system, their credentials are also used to log in to Mascot Server, so you don't need to enter a password.
You can read about how to implement these approaches in our recent blog article.
|
 |
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Quantifying the Impact of the Peptide Identification Framework on the Results of Fast Photochemical Oxidation of Protein Analysis
Marek Zakopcanik, Daniel Kavan, Petr Novak, and Dmitry S. Loginov
J. Proteome Res. 2024, 23, 2, 609–617
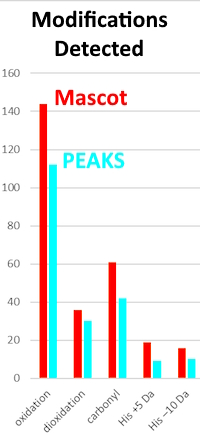
The authors undertook a detailed investigation of the performance of Mascot and PEAKS search engines for the identification of peptides and modification sites for a hemoglobin−haptoglobin Fast Photochemical Oxidation of Protein (FPOP) timsTOF data set.
As one of many examples, they cite Met15 oxidation in the peptide VADALTNAVAHVDDMPNALSALSDLHAHK in its triply charged form. Both search engines were able to identify numerous fragmentation ions and, thus, assign a very high score, but PEAKS did not account for relatively strong y14 and b18 ion signals assigned by Mascot. Furthermore, Mascot evaluated the certainty of oxidation localization as almost 100%, while PEAKS, on the other hand, evaluated its certainty only as AScore 11.12, which is a low rank. In other examples Mascot showed the ability to assess confident site determination, even with lower PSM scores while PEAKS AScore values do not provide reliable guidance on the certainty of modification localization.
In closing the authors state: We conclude that using Mascot for FPOP data leads to more reliable and understandable IDs and requires less manual validation.
|
|
|
|
 |
|
|
|
Mascot for your research lab
Many free software solutions exist for protein identification and protein quantitation. Some are open source; others are closed source but distributed as freeware. Why pay money for Mascot?
Here are a few reasons to get started:
- Mascot is mature software, 'battle hardened' under real-life use over 25 years and continues to be cited by over 2000 publications every year
- Excellent performance and scalability
- Database searches are fully reproducible
- Drill down into the data and inspect the evidence for yourself
- Extensive online documentation and training materials
- All aspects of processing are configurable and automatable
- Premium Support means you never have to diagnose issues on your own
If you need more reasons to put Mascot in your lab, read more in Mascot for researchers, which elaborates all the above points.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|