|
To view this email as a web page, click here. |
|
|
|
Welcome
We are looking forward to seeing many of you at the upcoming ASMS meeting in Anaheim, California. Sign up for the Matrix Science User Meeting on 3 June, and meet us at booth #219.
Are you presenting a poster or talk at ASMS that uses Mascot? Send us the poster/talk number, presenter name and abstract to newsletter@matrixscience.com and we will feature it in a future newsletter.
In this month's highlighted publication, an artificial neural network is used to optimize a sample preparation workflow.
We have been providing free public access to Mascot Server for 25 years.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Matrix Science at ASMS
|
|
|
See the latest Mascot developments this June – stop by at our booth #219 at the ASMS conference in Anaheim, California.
And join us at our annual ASMS User Meeting held on 3 June. There is no charge for attending the meeting, but advance registration is required. Numbers will be limited, so early registration is advisable. Breakfast will be provided.
Monday 3 June, 7:00 am – 8:00 am
Room 213A, Anaheim Convention Center, Anaheim, CA
Preview of Mascot Server 2.9,
Ville Koskinen, Matrix Science Custom reporting with Mascot Distiller,
Patrick Emery, Matrix Science Automatically running reports with the Mascot Daemon Export Extender,
Richard Jacob, Matrix Science
Register now
|
 |
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
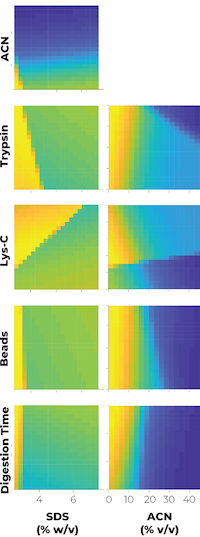
Refinement of paramagnetic bead-based digestion protocol for automatic sample preparation using an artificial neural network
Sergio Ciordia, Fatima Milhano Santos, Joao M.L. Dias, Jose Ramon Lamas, Alberto Paradela, Gloria Alvarez-Sola, Matías A. Avila, Fernando Corrales
Talanta 274, 1 July 2024, 125988
The authors investigated six factors in the single-pot solid-phase-enhanced sample preparation approach that affect protein extraction and digestion efficiency for MS analysis.
The goal was to maximize the number of peptides identified without missed cleavages in the HeLa proteome as a function of trypsin and Lys-C concentrations, percentages of ACN in the digestion buffer, digestion times, as well as the quantity of beads and the percentage of SDS.
They ran 30 experiments encompassing a range of these factors, and used these to train an artificial neural network model (ANN) to find the optimal conditions to maximize peptide IDs.
The ANN was able to predict optimal parameters without having to run an experiment for every factorial combination, while reducing tedious and laborious manual steps.
The best conditions for the digestion of 50 μg of HeLa extract are 2.5% SDS, 300 μg of beads, and long-term digestion (16h) with 0.15 μg Lys-C and 2.5 μg trypsin. These conditions were then adapted to the OT-2 automation platform where the number of peptide IDs with no missed cleavages were 36,614 and 36,708 using the automatic and manual workflows, respectively. The automatic protocol showed great reproducibility and low variability with additional sample types, including human plasma, A. thaliana leaves, E. coli cells and mouse tissue cortex.
|
|
|
|
 |
|
|
|
25 years of the free Mascot service
|
|
|
We have been providing a public Mascot service free of charge for 25 years.
Tens of thousands of people have finished over 27 million database searches (and counting!) since 1999.
We're able to provide the service for free thanks to several features we built into Mascot Server:
Designed for remote use – Search submission, reports and most admin tasks are done through the web browser. Fully automated – Once the system is set up, little manual admin is required. Runs on commodity hardware – Any standard Intel/AMD server is suitable. Few system dependencies – Use any Windows or Linux version. Fault tolerance – Mascot copes with invalid, faulty and incomplete input. Mascot Security – Automatically apply limits on the size and number of concurrent guest searches to ensure everyone gets a fair slice.
Additionally, we always run the most recent retail version of Mascot Server so you can have confidence that your results are the best you can expect.
Of course, if you want to automate search submission, perform large searches, search additional sequence databases, or customize the configuration, you would need to purchase an in-house license.
We hope you continue to enjoy this free service.
Read more in our blog, including news about a recent hardware upgrade.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|