|
To view this email as a web page, click here. |
|
|
|
Welcome
Using predicted retention times, Mascot Server 3.0 identifies more peptides than any previous version. A new tutorial shows how to use it.
In this month's highlighted publication, the authors reveal N-terminal methylation of Physcomitrella organellar proteins.
Updating to Mascot Server 3.0 is straightforward, with only a few known issues affecting third-party tools.
And an overview of new features and improvements in Mascot Server 3.0 is now available on Youtube.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
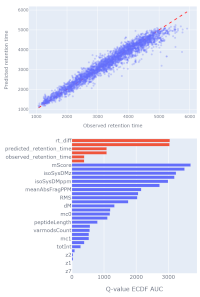
Selecting the best DeepLC model
|
|
|
Mascot Server 3.0 includes DeepLC, which is a retention time (RT) predictor that uses a convolutional neural network (deep learning) architecture.
The goal of RT prediction in Mascot is to increase the sensitivity of database search results.
Start by selecting a DeepLC model suitable for your column type and experimental parameters.
Mascot calculates the delta between the observed and predicted RT and combines it with core features.
Then, Percolator uses semi-supervised machine learning to find the optimal separation between correct and incorrect matches, based on the core features and RT delta.
Predicted RT is potentially very powerful, as it is 'orthogonal' to metrics calculated from the MS/MS spectrum.
Mascot ships with 20 models for different column and experiment types, but there is no need to try every one of them in turn.
The procedure for creating a short list is described in the tutorial.
Once you have found a suitable model for your experimental conditions, the procedure does not need to be repeated.
Just save the model in the Daemon parameter editor as the default, or save it as a cookie as a search form default.
Mascot takes the guesswork out of machine learning by providing a machine learning quality report.
The report includes metrics and visualisations for DeepLC model performance, which helps you quickly assess whether the chosen model is suitable for your data.
The full tutorial is available on our website.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
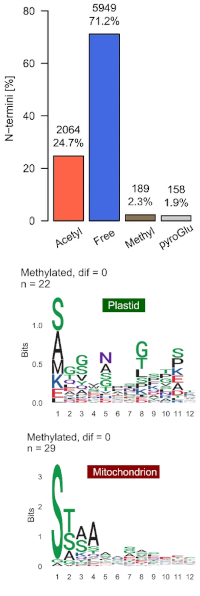
A snapshot of the Physcomitrella N-terminome reveals N-terminal methylation of organellar proteins
Sebastian N. W. Hoernstein, Andreas Schlosser, Kathrin Fiedler, Nico van Gessel, Gabor L. Igloi, Daniel Lang & Ralf Reski
Plant Cell Reports, 43, 250 (2024)
The authors used proteomic data from 24 different experiments to investigate N-terminal acetylation and methylation in Physcomitrella.
The Physcomitrella moss is a versatile model with broad application in basic research but also as a production platform for recombinant biopharmaceuticals.
The 24 experiments cover a range of tissues and treatment types.
In each experiment, N-terminal peptides were enriched, free amino groups blocked by reductive dimethylation and internal peptides were depleted after proteolysis.
After LC-MS/MS analysis, the spectra were searched with Mascot and imported in Scaffold 5.
The authors identified 11,533 protein N-termini corresponding to 4517 proteins at a strict 0.08% peptide FDR and 0.4% protein FDR.
Approximately 25% of identified N-termini were acetylated and 4% were methylated or carried a pyro-glutamate.
These new data can be used for optimizing computational targeting predictors and targeted localization of customized protein fusions.
An unexpected discovery is N-terminal monomethylation of plastid and mitochondrial proteins, affecting N-terminal alanine and serine as well as methionine.
The authors propose PpNTM1 (P. patens alpha N-terminal protein methyltransferase 1) as a candidate for protein methylation in plastids, mitochondria and the cytosol.
|
|
|
|
 |
|
|
|
Mascot Server 3.0 compatibility
|
|
|
Updating to Mascot Server 3.0 is straightforward: simply download the installer package and run setup64.exe (Windows) or extract the files over your existing installation (Linux).
We have worked hard to maintain backwards compatibility with application programming interfaces (APIs) and existing search results.
There are a few known issues with third-party applications, all of which have a workaround available.
Proteome Discoverer (Thermo Scientific): The Mascot node makes a version check, which fails because it expects major version number to be 2.x, not 3.x.
A hotfix is available on our website, and we have been advised by Thermo that the bug will be fixed in the next version of Proteome Discoverer.
Scaffold (Proteome Software): Current versions of Scaffold only support results in the old (.dat) format.
Please either export the results as "Mascot DAT" from the Mascot results report, or enable a compatibility setting (AlwaysCreateDat28ResultsFile) that forces Mascot to save results in the old format.
We are working with Proteome Software to enable support for MSR files in the next version of Scaffold.
BioTools and BioPharma Compass (Bruker): These products are able to submit searches but initially fail to download the results.
Please enable a compatiblity setting (AlwaysCreateDat28ResultsFile) that forces Mascot to save results in the old (.dat) format.
We are in contact with Bruker developers to help them solve the issue in BioTools and BioPharma Compass.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|