|
To view this email as a web page, click here. |
|
|
|
Welcome
Mascot Server 3.0 boosts peptide identifications with predicted fragment intensities – new tutorial is now available.
This month's highlighted publication illuminates the role of the self-ligating surface receptor on activated cytotoxic T-cells.
When you just need a simple HTML document of search results, save it from Protein Family Summary.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Selecting the best MS2PIP model
|
|
|
Mascot Server 3.0 includes MS2PIP, which predicts the MS/MS fragmentation spectrum from peptide sequence, charge state and modifications.
The goal of fragment intensity prediction in Mascot is to increase the sensitivity of database search results.
At the end of the database search, the predicted spectra of the search results are compared to the observed spectra.
For incorrect matches, the correlation between the predicted and observed spectrum is expected to be randomly distributed.
On the other hand, correct matches should have high correlation with predicted spectra.
Mascot combines the spectral correlation with core features and uses Percolator to separate incorrect matches from correct matches.
Mascot ships with 13 pre-trained models that cover most instrument types.
The CID model is a good starting point for all linear ion traps.
For Thermo Orbitrap instruments, the HCD2021 model is best.
The TTOF5600 model provides good predictions for Sciex 5600 and 6600 series instruments.
And the timsTOF2023 and timsTOF2024 models are suitable for Bruker timsTOF instruments.
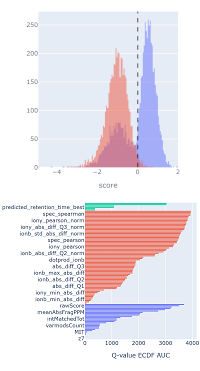
Mascot takes the guesswork out of machine learning by providing a machine learning quality report.
The report includes metrics and visualisations for MS2PIP model performance, which helps you quickly assess whether the chosen model is suitable for your data.
The full tutorial is available on our website, and a video tutorial is available on Youtube.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
SLAMF7 (CD319) on activated CD8+ T cells transduces environmental cues to initiate cytotoxic effector cell responses
Holger Lingel, Laura Fischer, Sven Remstedt, Benno Kuropka, Lars Philipsen, Irina Han, Jan-Erik Sander, Christian Freund, Aditya Arra & Monika C. Brunner-Weinzierl
Cell Death & Differentiation (2024), doi:10.1038/s41418-024-01399-y
The self-ligating surface receptor SLAMF7 is a clinically relevant target for novel cancer therapies.
The authors study the effect of SLAMF7 stimuli on CD8+ T-cells during the initiation of cytotoxic T-cell responses.
They identify a novel function, where CD8+ T-cells could utilise SLAMF7 to transduce environmental cues into cellular interactions.
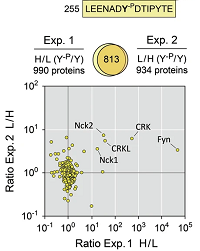
Pull-down assays and network analyses identified novel intracellular binding molecules involved in T-cell contact formation.
For the pull-down assay, CD8+ T-cells were lysed together with αCD28 antibodies plus phosphorylated and non-phosphorylated synthetic peptides.
Bound proteins were separated by SDS-PAGE, and digested and 18O-labeled in gel.
The peptides were analysed using LC-MS/MS in two replicates.
The 18O quantitation was performed by Mascot Distiller and database searching with Mascot Server.
|
|
|
|
 |
|
|
|
Saving search results as HTML
|
|
|
Mascot Server includes interactive reports for browsing database search results.
The results can be exported in various XML and spreadsheet formats, but for non-expert users, sometimes you just want a simple document of identified proteins and peptides.
It's easily done:
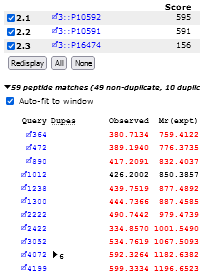
Open the search in the Protein Family Summary report, which is the default for MS/MS searches.
Select to view all protein hits on a single page, then click Expand All.
This expands the protein hit information and peptide match tables.
Save the page as HTML from the web browser and send it to the end user.
The HTML document contains the protein hit summary and the key peptide evidence supporting each protein hit.
If you don't need the peptide information, the Report Builder tab also makes it easy save a filtered table of protein hits as a CSV file.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|




|
|
|
|
|