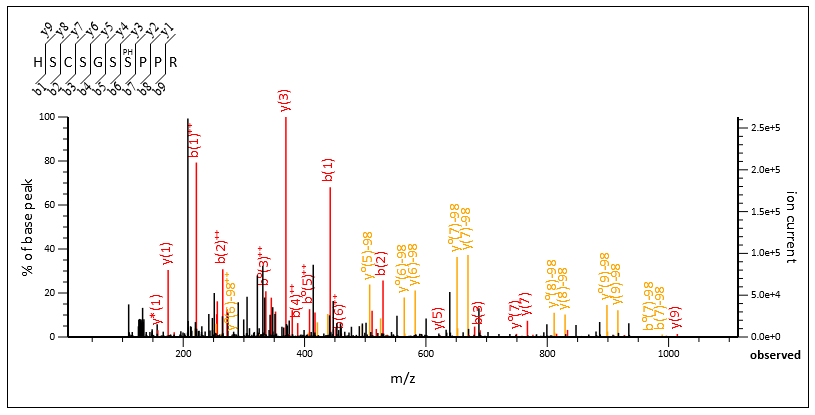
Proteomics solutions you can trust
Matrix Science develops software solutions for mass spectrometry proteomics. Mascot is a great choice for researchers, core labs, industry and educators.
Many third-party systems integrate directly with Mascot: Thermo Proteome Discoverer™, Bruker BioPharma Compass, Progenesis QI, Waters PLGS, Genedata Expressionist and Proteome Software Scaffold.
Blog
December 30, 2025
From time to time, a piece of software that has been working reliably suddenly doesn’t. The most common reason for Mascot Server to fail is running out of disk space, [...]
News
September 18, 2025
We are pleased to announce a new distributor for Matrix Science products! Customers in India and neighbouring countries can now buy Mascot Server through BioInnovations, who provide instruments, software, services [...]
Latest products
Protein identification
Mascot Server is the trusted reference standard for protein identification by mass spectrometry for 25 years. Mascot is designed for high throughput, and has no limits on the number of users, the size of the searches or the size of sequence databases. It continues to be cited by over 2000 publications every year.
Mascot Distiller provides additional tools for protein identification, including manual and automatic de novo sequencing.
Protein quantitation
Mascot Server includes native support for TMT/iTRAQ quantitation.
Mascot Distiller is a vendor independent Windows workstation application for quantifying proteins and peptides from a Mascot database search. Use any MS1 quantition, such as: SILAC, 18O, metabolic labelling and label-free methods (LFQ). Process raw files from Agilent, Bruker, SCIEX, Shimadzu, Thermo and Waters.
Batch automation
Mascot Daemon is a batch automation utility, bundled with Mascot Server. In combination with Mascot Distiller, every step of a quantitation experiment can be fully automated.
Mascot Parser provides an API (Application Programmer Interface) to Mascot Server result files and Mascot Distiller project files, suitable for implementing your own system integrations and pipelining.