Mascot Distiller – quantitation and more
Mascot Distiller Workstation is a vendor independent application for quantifying proteins and peptides from a Mascot database search. It offers a single, intuitive interface to native (“raw”) mass spectrometry data files from Agilent, Bruker, SCIEX, Shimadzu, Thermo and Waters.
Distiller capabilities
In addition to quantitation, Distiller includes a wide range of functionality for raw data processing and peptide identification. Several acquisition modes are supported:
- MALDI – MS (peptide mass fingerprinting) and MS/MS
- Data Dependent Acquisition (DDA) – MS/MS, LC-MS/MS, ddaPASEF
- Data Independent Acquisition (DIA) – Waters MSE (Thermo and Sciex: coming soon!)
Distiller’s capabilities increase with every major version:
- Native integration with Mascot Server for trusted and reliable protein identification
- Label-free quantitation, including retention time alignment (“match between runs”)
- Quantitate peptides and proteins with any MS1 method using a precursor mass shift (e.g. 18O, AQUA, ICAT, ICPL, Metabolic labelling, SILAC)
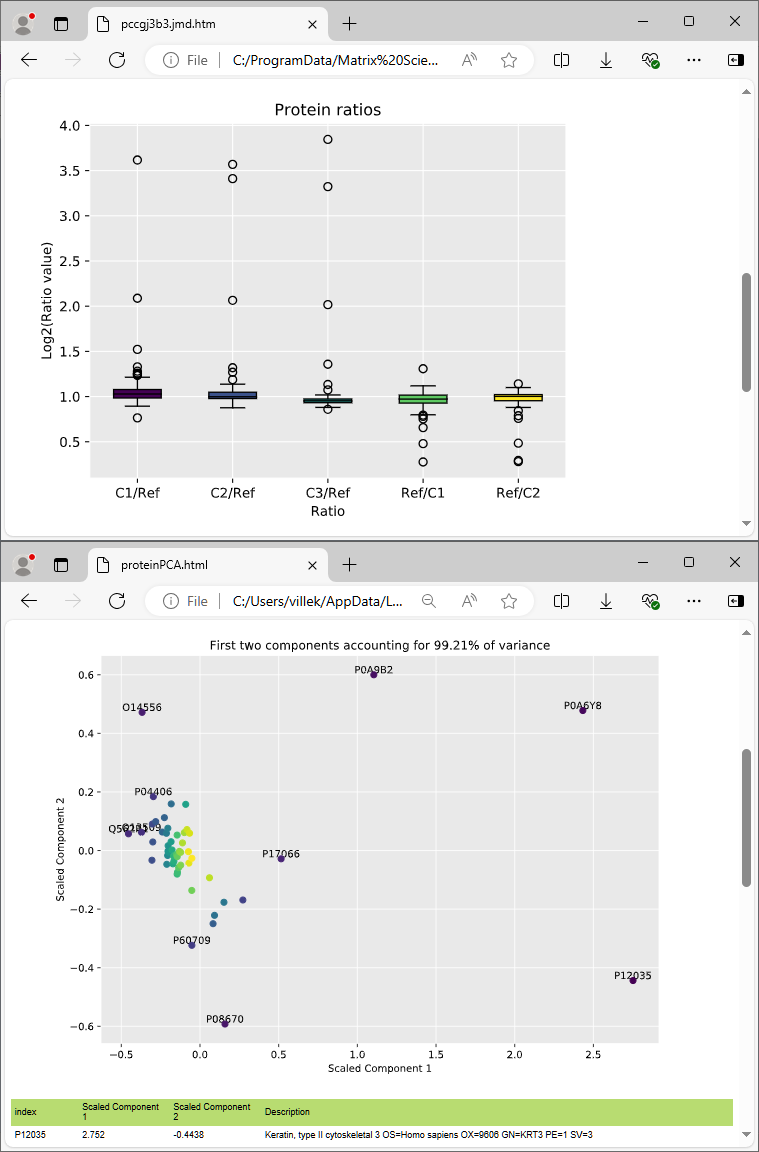
- Quantitation graphs and plots for publication and export results to statistical packages
- Augment database search results with de novo sequencing
- Improve peptide match quality with optimised peak detection
Claim a free 30-day product key, or get a quote today.
Supported file formats (DDA)
Mascot Distiller supports data dependent acquisition (DDA) from all major instruments used in proteomics research today, including:
- Agilent MassHunter (Q-TOF)
- Bruker TDF (timsTOF, ddaPASEF), baf (QTOF)
- SCIEX Analyst (5600, 6600, 7600 series)
- Thermo Xcalibur (LCQ, LTQ, Orbitrap)
- Waters MassLynx (QTof, M@ldi, TofSpec, Synapt)
- mzML 1.1 (XML interchange format)
- mzXML 2.0 and 2.1 (XML interchange format)
Mascot Distiller includes additional data access libraries for older instruments and peptide mass fingerprint (PMF) experiments. More details are available in the Distiller help supplied with the application.
Supported file formats (DIA)
Mascot Distiller supports data independent acquisition (DIA) experiments using:
- Waters MassLynx (MSE)
- Thermo: coming soon!
- SCIEX: coming soon!
In-house licence
Mascot Distiller is available for Microsoft Windows on Intel and AMD processors. The software is typically installed on a workstation computer at your own premises. The licence is perpetual, not an annual fee, and includes 1 year warranty covering technical support and all updates. After the first year, if you wish to continue receiving technical support and updates, you have the option of purchasing a support contract.
Mascot Distiller is shipped as a digital download. Get a quote, send us the purchase order and we will send you the download link.
Mascot Distiller integrates with Mascot Server for protein identification by database searching. We recommend the latest release of Mascot Server. Batch automation of quantitation requires Mascot Daemon, which is shipped as part of Mascot Server.
System requirements
Mascot Distiller requires 64-bit Microsoft Windows 7 SP1 or later, Server 2008 R2 SP1 or later. Distiller is fully compatible with Windows 11/Server 2022. In all cases, the latest service pack should be applied. Mascot Distiller requires .NET Framework 4.6 or later to run.
For PC hardware, we recommend an Intel Core i9 or AMD Ryzen 9 processor with at least 16 cores, 32GB RAM (preferably 64GB) and fast disk with at least 4TB of space. NVMe is beneficial but not required. Distiller does not use a GPU; all computation is done on the CPU.
Premium Support
A new licence includes one year of Premium Support, providing:
- Unlimited priority access to expert technical support
- Remote problem diagnosis and guided installation/migration
- Free update to any new release of the software
Your questions are answered directly by Mascot developers and experts based in the UK and USA.
You can continue receiving these benefits in subsequent years by purchasing an annual support contract. After the first year, if you wish to continue receiving technical support and updates, you have the option of purchasing a support contract for 35% of the licence fee per year.
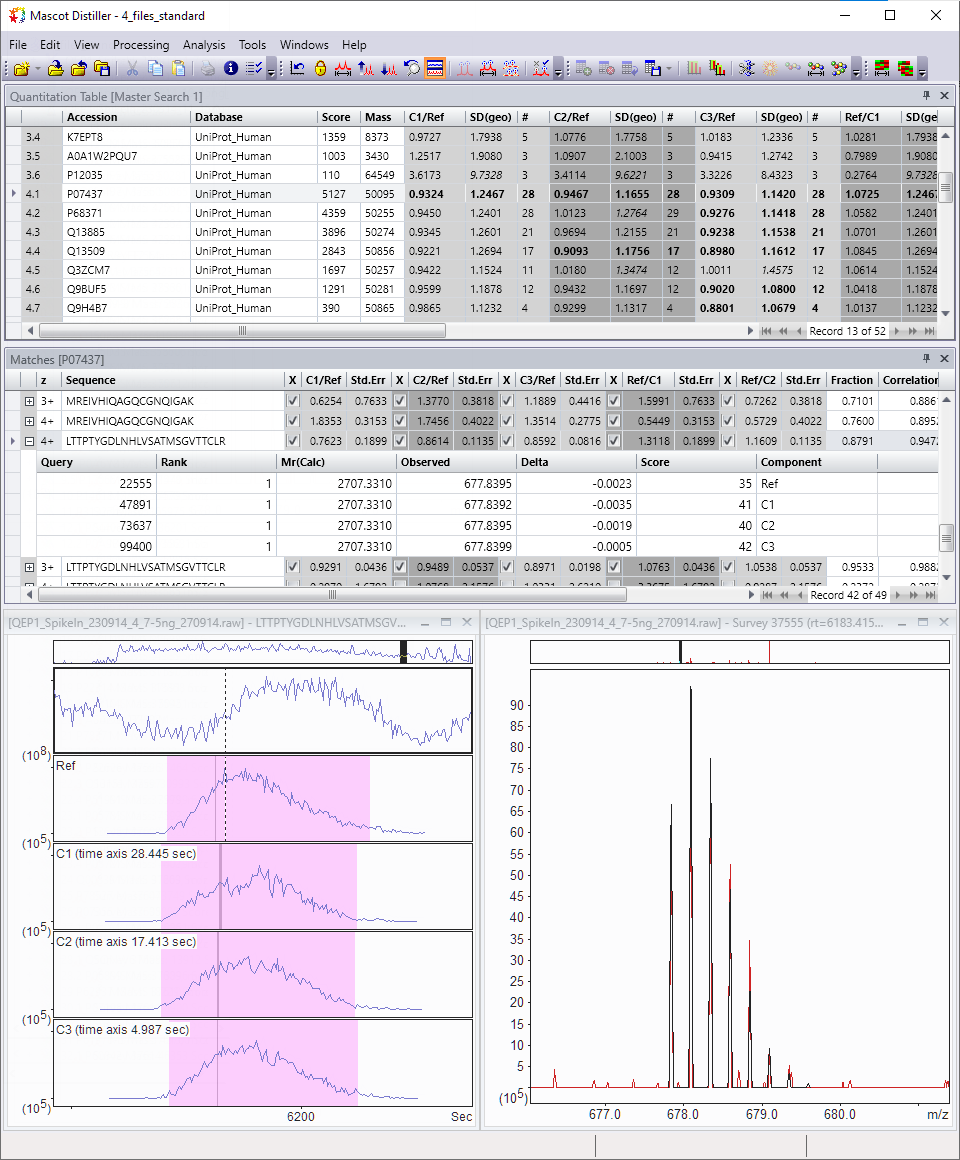
Quantitation
Distiller implements quantitation based on the relative intensities of extracted ion chromatograms (XICs) for precursors in MS1 survey scans. This approach can be used with label-free quantitation (LFQ) and any chemistry that creates a precursor mass shift, for example, 18O, AQUA, ICAT, ICPL, Metabolic labelling, and SILAC.
Quantitation results can be visualised and exported in a variety of interactive graphs, HTML reports and CSV spreadsheets.
Examples:
- Label-free quantitation in Mascot Distiller
- Fractionated label-free quantitation in Mascot Distiller
- Reporting quantitation datasets with Mascot Distiller
Batch automation with Mascot Daemon
The Mascot Daemon Toolbox enables the Mascot Distiller libraries to be called from Mascot Daemon, to process batches of data files and submit searches to a Mascot server. Peak lists are saved to disk automatically, and Distiller project files can also be created, allowing the raw data and peak list(s) to be opened in Distiller by clicking on a hyperlink in Daemon. Daemon saves the database search results in the Distiller project file. All stages of a quantitation experiment can be batch automated, form peak picking to saving the Distiller project complete with a quantitation report.
Search Toolbox
The Search Toolbox is a collection of tools for protein identification and characterisation:
- Display Mascot search results within Mascot Distiller
- Automatic de novo sequencing
- Automatic sequence tag calling
- Manual de novo and sequence tag
- Predict fragment ions from a peptide sequence and overlay them on an MS/MS spectrum
- Predict mass values from a protein digest and overlay them on a spectrum
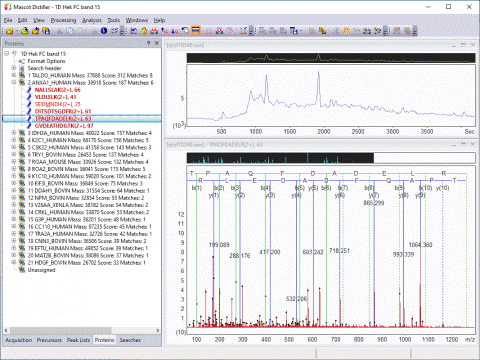
In the case of an MS/MS search, the top ranking peptide matches for each peak list are displayed under a Mascot Search Results node in DataSet Explorer. Each match is labelled with the peptide sequence and Mascot ions score.
The Search Toolbox includes a powerful de novo sequencing algorithm:
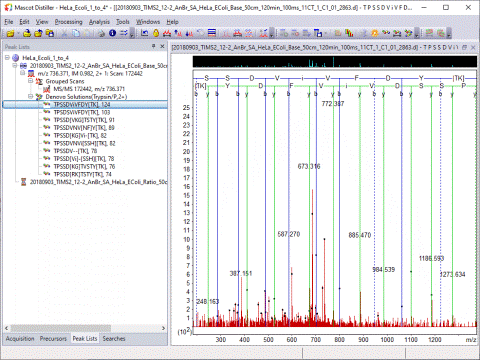
Optimised Peak Detection
Mascot Distiller includes a unique peak picking algorithm, which fits each peak to a complete isotope distribution. Raw data from any instrument is processed into high quality, de-isotoped peak lists.
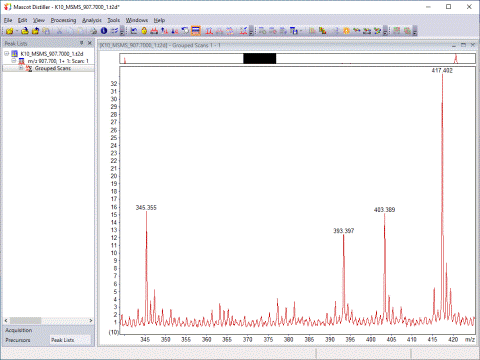
Mascot Distiller detects peaks by attempting to fit an ideal isotopic distribution to the experimental data. This ideal distribution is predicted from the elemental composition expected for a peptide of average amino acid composition at that point on the mass scale. The profile of the ideal distribution is then adjusted by varying the mass, resolution, intensity, and charge state, so as to maximise the correlation with the experimental data. Once the best fit has been obtained, the peak is added to the peak list and the corresponding signal subtracted from the spectrum. The process is repeated until all the significant peaks have been fitted.
The advantage of this approach is that the complete experimental isotopic distribution is fitted, not just the 12C peak. The charge state is automatically determined and the peak list contains only monoisotopic peaks, even when the signal to noise is poor or the isotopic distribution not fully resolved. Smoothing is not necessary or desirable.
Peak lists can be viewed, sorted, edited and saved to MGF format files, as well as a comprehensive format that includes additional information for each peak. Peak lists can be submitted direct to a Mascot server for database searching. The entire Mascot Distiller workspace, including peak lists and preferences, can be saved to a project file.