Complementary reporter ion clusters in TMT/TMTpro labeling
We recently received a support request about complementary ions in TMTpro labeling. Complementary ions are formed during fragmentation, where the precursor loses the reporter ion and carbon monoxide, leaving behind the peptide and the balance region of the label. Complementary ion spacing is similar to reporter ions but the mass varies depending on peptide mass. The customer was concerned that this might affect the peptide match score.
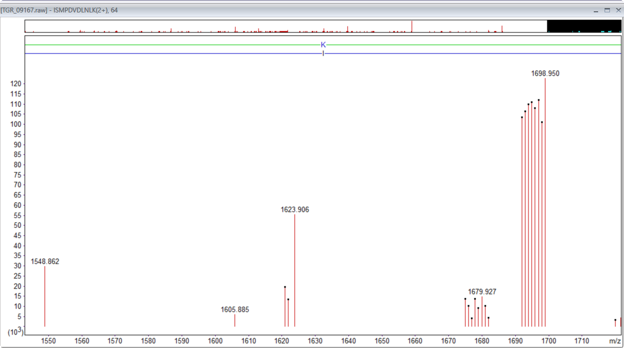
Figure 1: An example of the TMTpro singly charged complementary ions for the peptide ISMPDVNLNLK, m/z 926.546, 2+, Mr(measured) 1851.0775. The main complementary ions are in the region 1673 to 1698. There is a second lower intensity set that corresponds to the loss of water -18Da.
We wrote a script to process the spectra and remove the complementary ions to see if there was a change in the peptide score. In the customer’s data set, about 15% of the peptides were affected. After removing complementary TMTpro peaks, the score of the affected peptides increased by 3-4 on average. A small number of peptides, 124, scored lower after removal of the complementary ion region. These peptides were either misidentifying one of the complementary ions as a peptide fragment ion or there was an actual fragment ion in the same region as the complementary ions and it was removed.
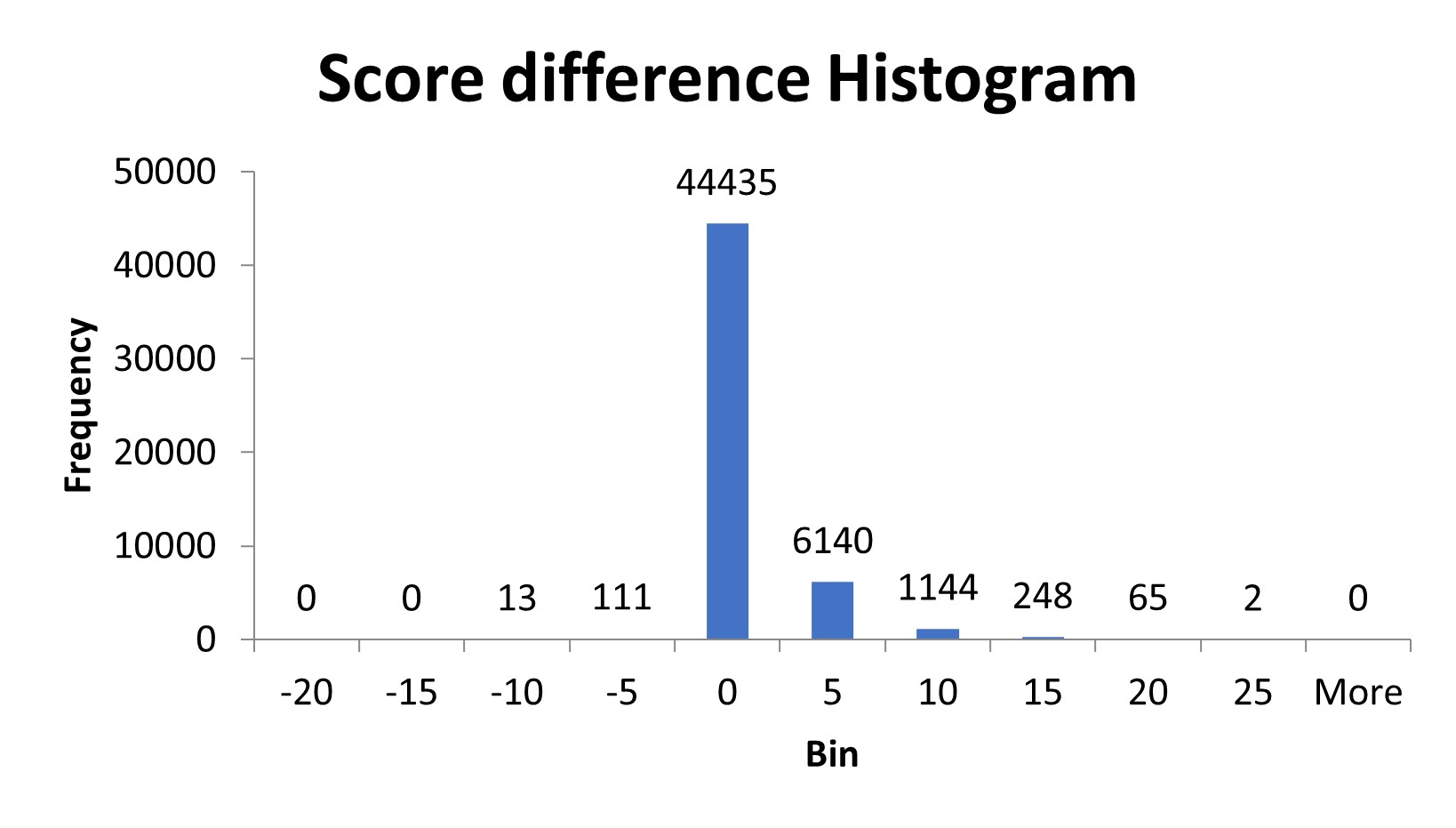
Figure 2: Histogram of the difference in peptide score after removal of complement ions from the spectra.
Rather than removing complementary ions, can we use them for quantitation? Their presence depends on the fragmentation energy used in the acquisition method. Ideally, you want to apply enough energy to break the balance portion of the label from the peptide and fully fragment the peptide, so that no complementary ions are present. Alternatively, you can set up a method that promotes the creation of the complementary ions. Wühr et al.1 and Sonnett et al.2 showed that TMT complementary ions can be used for quantitation. A recent paper by Johnson et al.3 expanded this method to TMTpro labels and termed TMTproC.
The main reason to use complementary ions for quantitation is that there can be interference in the standard reporter ion region from co-isolating peptides that then fragment along with the target peptide and distort the reporter ion measurements. The resolution of the mass spectrometer at higher masses means that it is not normally possible to separate the C and N isotope forms of the complementary ions used in the full TMT or TMTpro range, but a subset is still available, 5 channels for TMT and 8 channels for TMTpro.
When reviewing the data, we also observed that a lot of the heavier triply charged ions had doubly charged complementary ions.
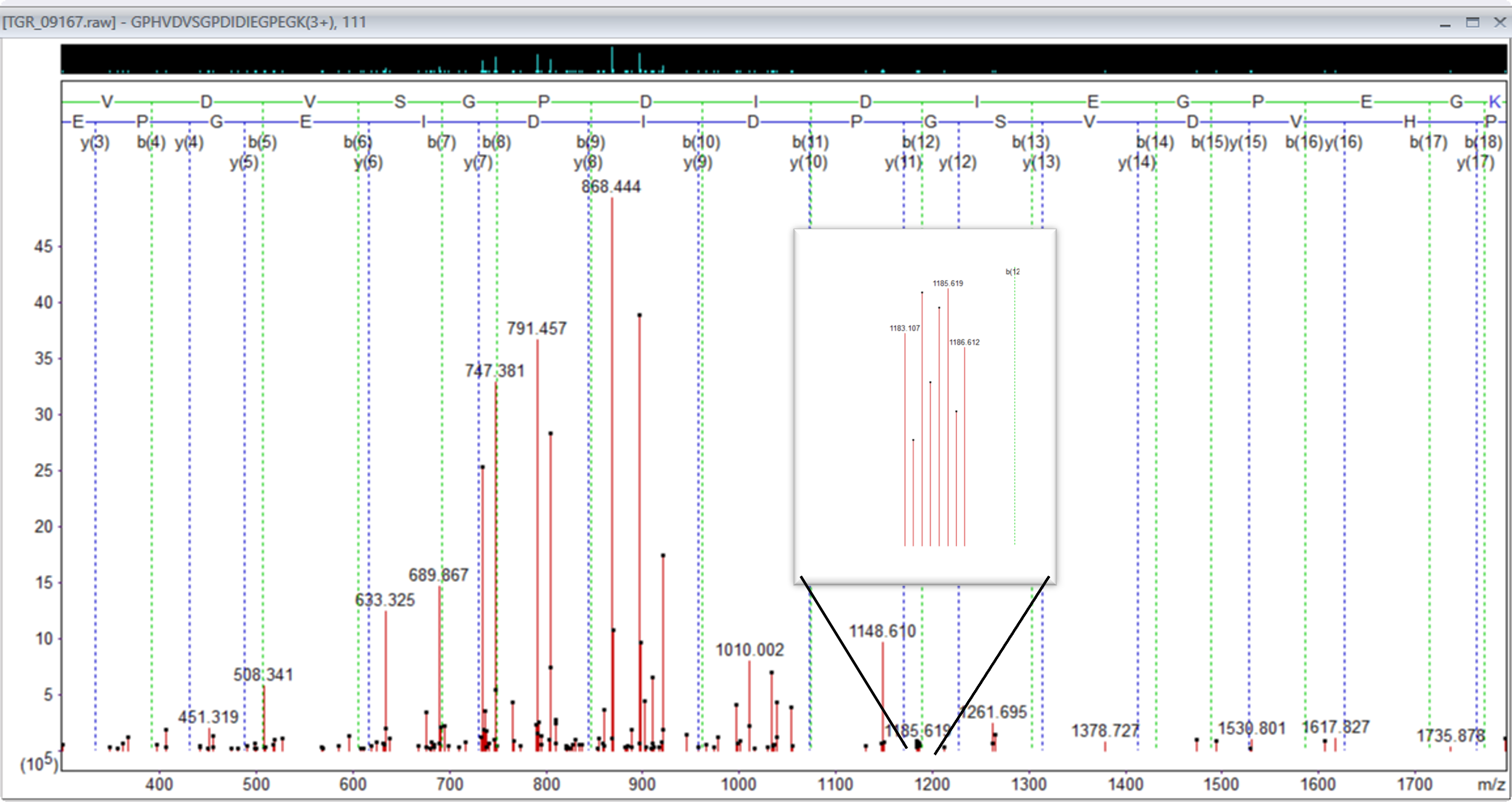
Figure 3: Precursor 842.781280,3+, showing doubly charged complementary ion region 1183 to 1186
We extended the script so that it can convert complementary ion masses into reporter ion masses and replace the original reporter ions in the spectra prior to searching and quantitation. The complementary ion regions, singly and doubly charged, are calculated from the precursor mass and the ions removed from the peak list along with the original reporter ion region. The spectra tend to contain only singly or doubly charged complementary ions, so the region with the most peaks was used to calculate the corresponding reporter ion masses. The masses were sorted lowest to highest and inserted back into the reporter ion region of the spectra. In this way we can use the existing Mascot Server quantitation methods to quantitate the results.
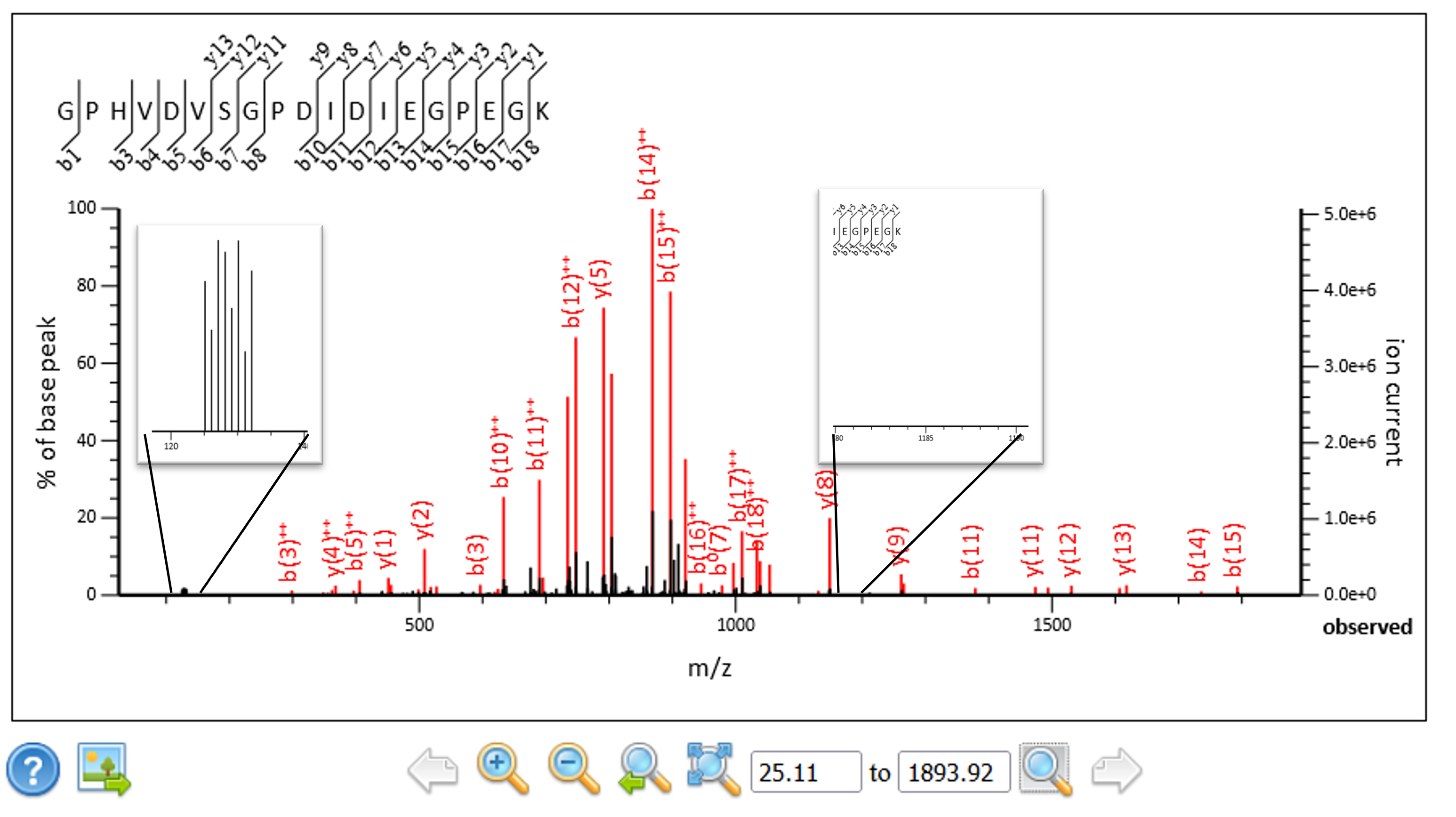
Figure 4: The complementary ions have been extracted from their original location and transformed into reporter ions.
A fractionated data set consisting of 24 files from the Johnson et al.3 publication was analyzed using the TMTproC method. The data is from a mixed sample with human cell lysate in 8 channels, all at 1:1:1:1:1:1:1:1 concentration, and yeast cell lysate in 6 channels at 0:1:5:10:10:5:1:0 concentration. The two labeled lysate samples are then mixed at a ratio of 10:1 Human:Yeast. The aim was to have a sample that has small but significant changes (yeast) in a background of unchanging (human) proteins. The experiment evaluates the signals from the yeast samples and looks for interference from the human samples. The Johnson et al.3 publication includes unfractionated comparisons of traditional MS2 reporter ion quantitation, MS3 reporter ion acquisition and TMTproC. In this analysis, we looked at the 24 file data set and evaluated the quantitation results in the same way as the publication. The following table shows the labels that were used and the transformed complementary ions for the TMTproC analysis.
| Label | Human | Transformed mass |
Yeast |
|---|---|---|---|
| 126 | 1 | 133 | |
| 127C | 133 | ||
| 127N | 132 | ||
| 128N | 132 | ||
| 128C | 1 | 132 | 1 |
| 129N | 1 | 131 | 5 |
| 129C | 131 | ||
| 130N | 130 | ||
| 130C | 1 | 130 | 10 |
| 131N | 1 | 129 | 10 |
| 131C | 1 | 128 | 5 |
| 132N | 127 | ||
| 132C | 127 | ||
| 133C | 1 | 127 | 1 |
| 133N | 126 | ||
| 134N | 1 | 126 |
A database search found ~37,600 peptides matching yeast proteins from ~317k total peptide matches at 1% FDR. No additional filtering was used on the yeast peptides beyond the default quantitation method quality control. The yeast set includes peptides from protein families that contain both human and yeast members. We can see from the histograms how the ratios reported for the peptides matching to a yeast protein are close to the expected ratios of 5:1, 10:1 and 10:5 with a bit of broadening from the interference of the human peptides. Filtering out the peptides from protein families with mixed human and yeast members may improve the histograms, but it’s not something you could do when looking at small changes in a homogeneous sample.
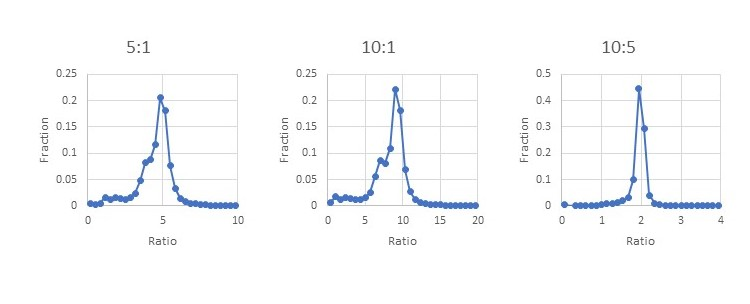
Figure 5: Evaluating ratio distortion for TMTProC in the 24 fraction data set.
In conclusion, if your TMT/TMTpro data acquisition method produces some unwanted complementary ions, then removing them prior to searching is slightly beneficial to the scoring of the peptides. Converting them into reporter ions allows Mascot Server to quantitate peptides using TMTc and TMTproC protocols. You can download the script, TMT_complementary_ions.pl, from our our help page, which also contains usage instructions and automating the preprocessing using Mascot Daemon. The script has an additional option to use the doubly charged complementary ions that is not mentioned in the original description of the TMTproC quantitation method.
Full support for complementary ion quantitation without the use of an intermediary script will be added to a future release of Mascot Distiller.
References
1 Wühr M, Haas W, McAlister GC, et al. Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Anal Chem. 2012;84(21):9214-9221. DOI: 10.1021/ac301962s
2 Sonnett M, Yeung E, Wühr M. Accurate, Sensitive, and Precise Multiplexed Proteomics using the Complement Reporter Ion Cluster. Anal Chem. 2018; 90(8):5032-5039. DOI: 10.1021/acs.analchem.7b04713
3 Johnson A, Stadlmeier, M, Wühr M. TMTPro Complementary Ion Quantification Increases Plexing and Sensitivity for Accurate Multiplexed Proteomics at the MS2 Level. J. Proteome Res. 2021; 20(6):3043–3052. DOI: 10.1021/acs.jproteome.0c00813
Keywords: Mascot Daemon, Mascot Distiller, quantitation, reporter, scoring, TMT