Matching and scoring internal fragments
Mascot ships with several instrument definitions, which define the fragment ion series used for matching and scoring. All of them enable b ions and most enable y ions, and a few like ETD-TRAP also enable c and z+1. Mascot can also match internal fragments, which are formed by double backbone cleavage, a combination of a/b type and y type. Of the default instruments, only MALDI-TOF-TOF and MALDI-TOF-PSD have internals enabled. However, enabling it for HCD can make a substantial improvement, especially with stepped collision energies.
This example is courtesy of Darryl Pappin, Cold Spring Harbour Laboratory. The run is a standard QC injection of HeLa on Thermo Exploris 480, using stepped collision energy mode.
Data was collected using a three-step regime with HCD collision energy (%) of 30, 35 and 40. When we first got the Orbitrap Lumos in 2016 we tended to use collision energies on the higher end of the normal range for HCD. This gave really good results for larger peptides and some PTMs. Phosphopeptides were great, as pretty much all the ion series you saw were the -80 or -98 neutral loss rather than a mix of fragments. One thing we also noticed fairly quickly was that the higher collision energies were giving significant numbers of internal fragment ions.
For the last couple of years we have been using this 3-stepped collision regime. The overhead for doing things this way is only about 5% more than the time for a single MS2, so it is pretty efficient. However, the fragments are now split between the energy levels, so peaks are a bit smaller than if you hit it with just one energy. However, it produces a good mix of internals for you to see.
Darryl also reports that internal fragment ions have been useful in crosslinking studies, where overall fragmentation is often sparse, and sometimes internals provide the only good evidence for one or the other of the two crosslinked chains.
The QC injection has 81,267 queries, acquired in high-resolution (FT) MS1 and MS2. Peak picking was done in Mascot Distiller using the default Thermo options. Precursor tolerance is set to 5ppm and fragment tolerance to 10ppm, as the instrument is very well calibrated.
The table below summarises the effect of enabling internals. Simply enabling ya and yb series gives 16% more matches at 1% PSM FDR and 13% more peptide sequences.
| Search | Target PSMs | PSM FDR | Target sequences |
Seq. FDR |
|---|---|---|---|---|
| ESI-TRAP | 13632 | 1.00% | 4355 | 0.90% |
| ESI-TRAP with internals | 15874 | 1.00% | 4918 | 1.10% |
The peak lists and search space are the same in both cases, so the improvement comes exclusively from higher peptide scores. Many matches that were previously below threshold (not statistically significant) now get a score above threshold. This improves both the number of duplicate matches, which are repeat PSMs of the same peptide, and the number of unique peptides identified.
You might expect search duration to increase when internals are enabled. However, duration for this small search was about the same in both cases. If your search space has many long peptides, the search will probably take a bit longer with internals than without, but we’re talking about a few percent increase in search duration.
The reason for the score improvement is quite interesting. During peak matching, Mascot divides the MS/MS spectrum into 100Da windows and selects peaks in intensity rank order. These are matched to calculated fragments. If a tall peak fails to match any calculated mass, Mascot subtracts a penalty term proportional to the total unexplained intensity. When internals are disabled, any medium to high intensity peaks due to double cleavage go unexplained. This results in a score penalty. When internals are enabled, Mascot calculates their masses and matches them to observed peaks. This reduces the number of unexplained peaks, which reduces the penalty term and the final match score is increased.
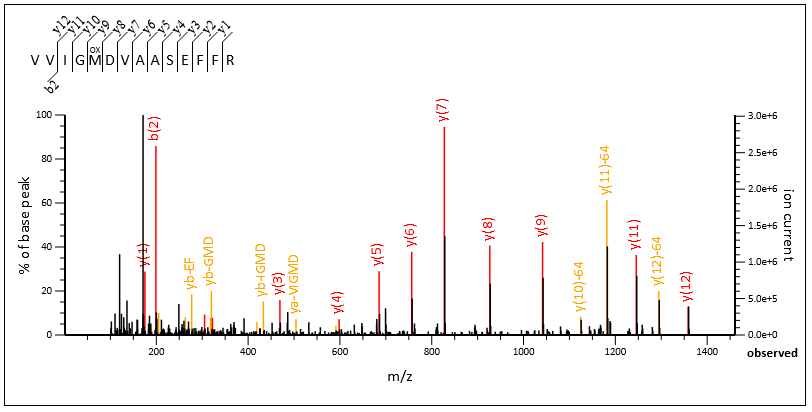
Query 57256, above, is a great example. The number of selected peaks is the same in both cases (“51 most intense peaks”). When internals are enabled, Mascot is able to label 13 extra peaks as internals, shown with yellow labels in Spectrum Viewer. When internals are disabled, these peaks are considered noise. Match score goes from 57 (disabled) to 71 (enabled).
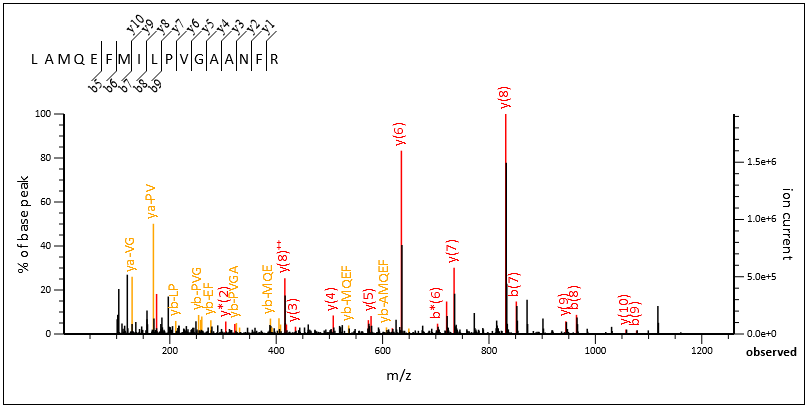
Query 70020, above, is another one. This time, enabling internals allows Mascot to go deeper into the spectrum. The algorithm finds not only 21 new double cleavage ions but also additional b and y peaks, which increases sequence coverage. The fairly high intensity of the double cleavage ions previously “masked” the weaker fragments.
Many more similar cases can be found. There are also a few cases, like query 56567, where the score does not change. As explained above, internals only contribute positively by reducing the noise penalty. They are otherwise ‘optional’ fragments, and their absence has no negative impact.
Finally, if you’re using Mascot through a third-party interface like Thermo Proteome Discoverer, change the instrument definition in the Mascot configuration editor, then select the right instrument in the search parameters. Mascot will use internals in matching and scoring, and nothing else needs to be changed for importing the results into the third-party tool.