Troubleshooting ML integration with Mascot client software
Getting started
Please follow the instructions for using machine learning with Mascot Server and specific client software:
- Using machine learning with Mascot Server 3.1 and Thermo Proteome Discoverer™ (4 pages, 253kB)
- Using machine learning in Mascot Server 3.1 with Mascot Distiller (5 pages, 224kB)
If the integration does not seem to work, follow the sections below for troubleshooting.
Did refining actually happen?
Check a log file to confirm Mascot successfully refined the results and sent them to the client software. (Not all client software programs have a mechanism to check with Mascot Server that refining has been done.)
1. Find the Mascot job number.
If you’re using Proteome Discoverer: open the the job queue. For example:
Mascot Info Mascot result on server (filename=../data/20241215/F007074.msr)
If you’re using Mascot Distiller: switch to the Searches tab in the tree view. Hover the mouse over the search results node, which displays the path and job number in a tooltip: for example, ../data/20241215/F007074.msr. Alternatively, right click and select Open Full Report in Browser, then look at the ‘file’ argument in the URL.
2. Check the API log file on the Mascot Server hard disk.
Open the log file mascot/logs/workarounds/client_result_file_mime_refining.log on the Mascot Server hard disk. Confirm that the refined results were sent for this job, for example:
[result_file_mime][8412][../data/20241215/F007074.msr] Refining the results.
[result_file_mime][8412][../data/20241215/F007074.msr] Refining succeeded.
[result_file_mime][8412][../data/20241215/F007074.msr] Preparing to combine '../data/20241215/F007074.msr'
with target and decoy pop files.
[result_file_mime][8412][../data/20241215/F007074.msr] About to run combine_dat28_with_pop.pl
[result_file_mime][8412][../data/20241215/F007074.msr] Successfully created MIME format file
..\data\cache\2024\12\wpjgy6beq7xy56bwbti7nokmmq\refined.dat.292179
[result_file_mime][8412][../data/20241215/F007074.msr] Dumped refined data in MIME format to standard
output. Done.
If the log file indicates that no refining was done, follow the steps below.
Test the instrument definition in Mascot
- Go to your local Mascot home page and Access Mascot Server. Select the MS/MS Ions Search.
- Select a typical MGF file, and select SwissProt as the database.
Choose the instrument you added (e.g. MS2PIP:HCD2021) and disable refining results with machine learning in the search form, as shown in the below screenshot:
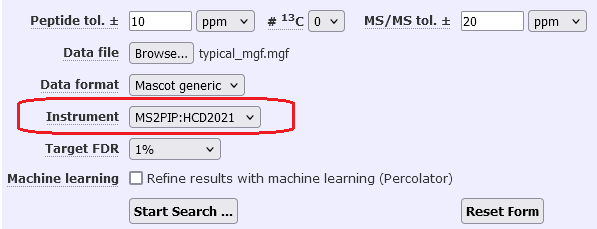
- Submit the search.
When the search parameters are set this way, it simulates how the search is submitted from the client software. When the results report loads, it should default to refining the results with the MS2PIP model selected in the instrument definition. You can also perform this test by repeating an existing search from the Mascot search log.
Mascot option ClientResultFileMimeRefining
Go to your local Mascot home page and open Configuration Editor. Open Configuration Options.
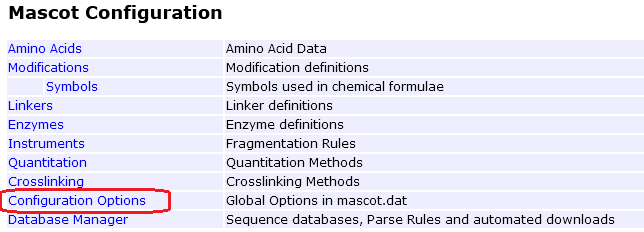
Check that the option ClientResultFileMimeRefining is present and enabled (1). This option is enabled by default in a fresh Mascot Server 3.1 installation. Updating to Mascot Server 3.1 should also add and enable the option.
If the option is present but set to 0, change it to 1.

Why are peptide scores different between the client software and Mascot Server?
A Mascot client software application imports two key values from Mascot Server for each peptide-spectrum match (PSM): the Mascot ions score and qmatch. The identity threshold is calculated from qmatch and the significance threshold. Then, the client software calculates the expect value from the score and the identity threshold. A PSM is significant if its expect value is below the significance threshold. Equivalently, a match is significant if its score is above the identity threshold.
When refining with machine learning is disabled, the client software displays the unmodified Mascot ions score and identity threshold. The identity threshold has a lower bound of 13, so any PSM with score below 13 can never be statistically significant, no matter how high you set the significance threshold.
When refining is enabled, Mascot sends a modified Mascot ions score and a modified qmatch to the client software. The modified score is -10log10(PEP) + 7, where PEP is the posterior error probability estimated by Percolator. The modified qmatch is qmatch=100, which forces the identity threshold of all PSMs to 20 at the default significance threshold (effectively the same offset, 13+7).
If you view the same results in the Protein Family Summary report in Mascot Server, you’ll notice that PSM score 13 is reported as score 20 in the client software. The reason for the offset +7 is to allow the FDR estimation algorithm to set the significance threshold to a value higher than 0.05. For example, to reach 1% FDR, the required PEP threshold may be 0.1256. This corresponds to the unmodified score -10log10(PEP) = 9. However, because the identity threshold has a lower bound of 13, a PSM with score 9 (PEP 0.1256) could never become statistically significant. This is why Mascot sends the modified score -10log10(PEP) + 7 = 16 and applies the same offset to the identity threshold. Now the client software is able to set a PEP threshold 0.1256 to reach 1% FDR.
Although the absolute value of the PSM score and identity threshold are different when viewing the results between the Protein Family Summary report in Mascot Server and the result viewer in the client software, the expect value of the PSM should be the same in both. For example, when refining with machine learning is enabled, a PSM with score 9 in Protein Family Summary has PEP 0.1256 and expect value 0.1256. The same PSM in the client software should have score 16 and expect value 0.1256.
