Mascot Distiller quantitates more proteins with machine learning
Mascot Distiller can now use machine learning with LFQ, TMT and other quantitation methods, thanks to API changes made in the recent Mascot Server 3.1 release. Machine learning typically increases the number of identified peptides at a fixed false discovery rate, which often leads to a substantial boost in the number of proteins quantified. We demonstrate this with an LFQ benchmark data set, where Distiller now quantitates 14% more proteins at 1% sequence FDR.
How it works
The changes made in Mascot 3.1 to enhance integration with Themo Proteome Discoverer™ are outlined in this blog article, and the same approach is used in Mascot Distiller – the current release of which, like Proteome Discoverer, lacks an interface for the new machine learning adapter settings.
Mascot Distiller 2.8.5.1 is a free update for Mascot Distiller which fixes a single bug in HTTP/S timeout handling and is required for the machine learning refined results to be imported into Distiller.
Briefly, you define a new instrument definition in Mascot 3.1 which specifies the MS2Rescore model(s) to automatically apply to the results. When the new definition is selected for a search, the refined results are returned to Mascot Distiller instead of the standard Mascot scores. This can give you a significant improvement in the number of proteins and peptides identified and quantified in any supported dataset.
Example: Thermo Orbitrap QE HF-X DDA run of human, yeast, E. coli mixture (PXD028735)
PRIDE project PXD028735 is raw data for “A comprehensive LFQ benchmark dataset on modern day acquisition strategies in proteomics (Pyuvelde et al., Scientific Data, 9(126), 2022)”. The authors used six instruments and six different mixtures of yeast, E. coli and human proteins. Every sample was run with every instrument with DDA.
We downloaded a set of three replicates (alpha, beta and gamma) for the spiked in samples (A and B) analysed on a Thermo Orbitrap QE HF-X. Samples A and B are comprised of mixtures of Human, S.cerevisiae and E.coli proteins in different amounts such that the expected ratio of a protein from Sample A / Sample B are as follows:
| Species | Expected Log2 protein ratio |
|---|---|
| H.sapiens | 0 |
| S.cerevisiae | 1 |
| E.coli | -2 |
In Mascot Server 3.1, set up a new instrument MS2PIP:HCD2021, where refining is enabled and the MS2PIP model HCD2021 is selected. The instructions for setting up the instrument can be found in Using machine learning in Mascot Server 3.1 with Mascot Distiller (5 pages, 225kB).
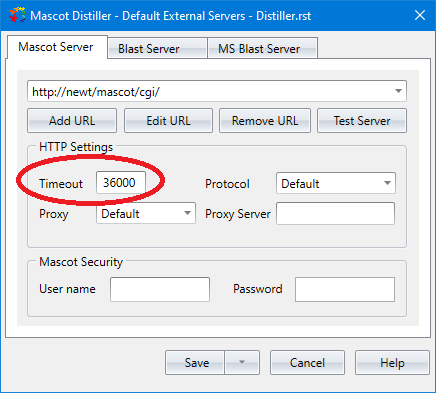
Once you have updated to Mascot Distiller 2.8.5.1, open the software and without creating a project, open the main Mascot Distiller workstation GUI and open the Tools->External Servers dialog. On the Mascot Server tab, under HTTP Settings, increase the Timeout value from the default of 60 seconds to a much higher number such as 36000 (10 hours). This is required because the connection between Mascot Distiller and the Mascot Server will have to be held open while the results are refined on the Mascot Server. During this period, no data will be sent from the Mascot Server to Mascot Distiller, and without increasing the timeout setting the connection will be terminated by Mascot Distiller before refining has completed.
Next, create a new multifile project in Distiller, selecting all six files and choose the default.ThermoXcalibur.opt processing options. When the project has opened, go to the Tools->Preferences->Mascot Search settings tab and use the following parameters:
- Protein Databases: Use human, yeast and E. coli Uniprot proteomes
- Enzyme Name: Trypsin/P
- Maximum Missed Cleavages: 2
- Instrument: MS2PIP:HCD2021
- Decoy: True
- Taxonomy: All entries
- Error Tolerant Search: False
- Precursor Mass Tolerance: 10 ppm
- Fragment Mass Tolerance: 20 ppm
- Variable Modifications: Oxidation (M), Actyl (Protein N-Term)
- Fixed Modification: Carbamidomethyl (C)
- Quantitation: Label-free [MD]
Mascot Distiller 2.8 is using an older version of the search form than the default used with Mascot 3.1. Therefore, it’s important to remember to select Decoy on the search form as this is required for the machine learning to run.
Now choose the “Process and Search” in Mascot Distiller – with the MS2PIP:HCD2021 option selected, the results will be automatically refined with machine learning and the refined results imported into Mascot Distiller. Once all the searches have been completed and merged, you can run quantitation on the results.
Over 25% more peptide matches using Machine Learning with MS2PIP:HCD2021
The table below summarises the protein and peptide counts with and without the HCD2021 model and Mascot Distiller 2.8.5.1:
| Mascot Server | #Protein hits | #Proteins quantified | #Peptide sequences (1% FDR) |
|---|---|---|---|
| 3.0 (no refining) | 5089 | 3235 | 27080 |
| 3.1 (with MS2PIP:HCD2021) | 5961 | 3699 | 34222 |
| %improvement | 17% | 14% | 26% |
With the HCD2021 model and machine learning enabled, we’re getting a 26% more peptide sequences identified, and 17% more protein hits – so we’re not just increase the coverage of already identified proteins and we’re also not just finding more one hit wonders.
More proteins quantified without loss of accuracy
After running label-free quantitation on the results, we can also see an improvement in the number of proteins quantified in two or more of the replicates by two or more peptide sequences while retaining very similar accuracy – the results are summarised in the table below:
| Mascot Server | Organism | Expected ratio (log2) | Median obs. ratio (log2) | Median absolute deviation | #proteins |
|---|---|---|---|---|---|
| 3.0 (no refining) | H.sapiens | 0 | -0.017 | 0.1 | 2369 |
| 3.1 (with MS2PIP:HCD2021) | H.sapiens | 0 | -0.012 | 0.1 | 2675 |
| 3.0 (no refining) | S.cerevisiae | 1 | 1.01 | 0.16 | 733 |
| 3.1 (with MS2PIP:HCD2021) | S.cerevisiae | 1 | 1.02 | 0.17 | 857 |
| 3.0 (no refining) | E.coli | -2 | -1.96 | 0.38 | 133 |
| 3.1 (with MS2PIP:HCD2021) | E.coli | -2 | -1.90 | 0.43 | 167 |
Median protein ratios for both refined and unrefined results are very similar, and close to the expected ratios for each organism. However, refining the results with MS2PIP:HCD2021 and Percolator has increased the number of proteins quantified with 2 or more peptides, from 3235 to 3699, so the increase in overall identifications is also observed in the label free quantitation results.
Keywords: label-free, machine learning, Mascot Distiller, MS2PIP, Percolator, quantitation