Searching DIA data, especially Swath
DIA-Umpire is a new, open source Java program that enables untargeted peptide and protein identification and quantitation using DIA data. A detailed description can be found in Nature Methods. The DIA-Umpire signal extraction module deconvolutes the DIA data to create a conventional DDA-type peak list, suitable for database searching. The software is intended to be applicable to DIA from any instrument, although the examples in the publication are mostly Swath on an AB Sciex 5600, with one example of data from a Q Exactive Plus.
The choice of mzXML as the input format is slightly unfortunate, as this became obsolete in 2008, when HUPO PSI released mzML. The suggested workflow for AB Sciex data is to convert Wiff to mzML using the AB Sciex MS Data Converter then convert mzML to mzXML using msconvert.
Example data files can be downloaded from SourceForge and more are available from Pride. Memory requirements mean using a 64-bit system for all but the smallest files. Processing LongSwath_UPS1_1ug_rep1.mzXML using the suggested parameters ups.se_params yielded three MGF files. Tier 1 (*_Q1.mgf) representing pseudo MS/MS spectra that are linked to high quality MS1 precursor features (3 or more detected isotope peaks), tier 2 (*_Q2.mgf) representing lower abundance precursors (2 detected isotope peaks only), and tier 3 (*_Q3.mgf) representing precursors detected in MS2 scans. All three peak lists were searched against human proteins in SwissProt:
| MGF | Results | # PSMs | # PepSeq | Unique PepSeq |
|---|---|---|---|---|
| LongSwath_UPS1_1ug_rep1_Q1.mgf | Tier 1 | 905 | 500 | 130 |
| LongSwath_UPS1_1ug_rep1_Q2.mgf | Tier 2 | 9 | 9 | 1 |
| LongSwath_UPS1_1ug_rep1_Q3.mgf | Tier 3 | 644 | 393 | 26 |
Counts are at 1% FDR and Unique PepSeq is the count of peptide sequences not found in either of the other tiers. For this particular analysis, almost nothing is found in tier 2. An area-proportional Venn diagram (eulerAPE) for counts of distinct peptide sequences at 1% FDR looks like this:
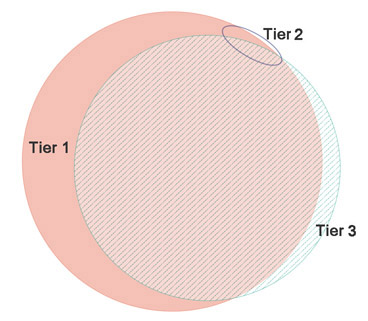
The DIA-Umpire documentation states that "These spectra are written to separate files, because they must be searched separately against a protein database as a consequence of differences in FDR estimates for these varying quality data." Using Mascot, the same search parameters can be used for all three, the score distributions for tier 1 and 3 are very similar, and the significance threshold for 1% FDR is essentially the same for tier 1 and 3, so no particular need for separate searches. If this picture applies generally, you might choose to search only the tier 1 peak list or, for best coverage, merge the tier 1 and 3 peak lists.
The file conversion requirements make automation tricky. Mascot Daemon 2.5 supports both AB Sciex MS Data Converter and msconvert, but not as multiple steps in a file conversion chain. If you are determined, you can set up four separate real-time monitor tasks, one to convert Wiff to mzML, a second to convert mzML to mzXML, a third to create MGF files from the mzXML using DIA-Umpire, and a final task to search each of the MGFs. This last task is necessary because DIA-Umpire doesn’t allow the output directory for the MGFs to be specified, so there has to be an additional task to pick up the MGFs from the same directory as the mzXML. Ignore the errors in the Daemon event log from the searches for the first three tasks, which are bound to fail.
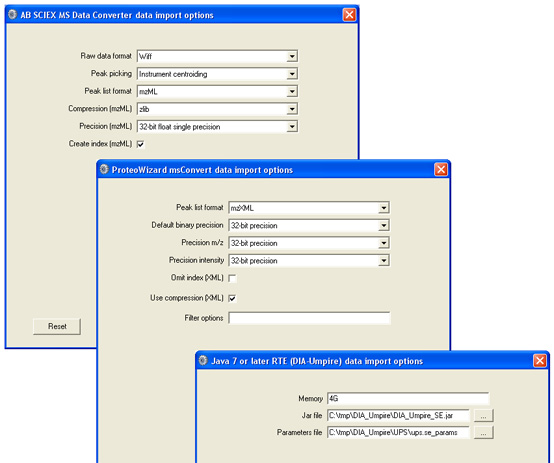
You can download an updated daemon_di_utils.xml, containing a definition for DIA-Umpire and with the mzXML output option added to the msconvert definition. Note that there is an error in the DIA-Umpire instructions: when using msconvert to transfrom mzML to mzXML, do not specify peak picking because this causes msconvert to throw an exception. The following settings seem to work:
Keywords: DIA, Mascot Daemon, SWATH
Hi John,
It’s a nice work on supporting DIA-Umpire on Mascot Daemon. I am very happy to work with you to streamline the process. And also supporting mzML is on the list but I really need to get time to fully test that. So let me know if there is something I could do at my side.
Update: I tried processing the same file (LongSwath_UPS1_1ug_rep1.mzXML) in the current release of DIA-Umpire (2.1.2). Results were not so good. PSM counts at 1% FDR using search conditions as above
ver 1.284 with ups.se_params: 905
ver 2.1.2 with diaumpire_se_ABSciex_params.txt: 326
ver 2.1.2 with ups.se_params: 394
DIA-Umpire Manual is still ver 1.4 and the support forum has no messages since July 2016, so I didn’t spend a lot of time investigating.