What’s new in Mascot Server 3.0
New versioning
Mascot Server 3.0 is a major release. From now on, a major release is indicated by the first component of the version number. The patch releases to Mascot Server 3.0 will have numbers like 3.1 and 3.2; then the next major version after this one will be 4.0.
Refine results with machine learning
Mascot Server 3.0 ships with Percolator, which is a semi-supervised algorithm for rescoring search results. Mascot Server now also ships with MS2Rescore 3.0, which is a “Modular and user-friendly platform for AI-assisted rescoring of peptide identifications” developed at the CompOmics lab at the University of Ghent.
Use predicted retention time (DeepLC) and predicted spectra (MS2PIP)
MS2Rescore includes DeepLC for retention time prediction and MS2PIP for predicting MS2 fragmentation spectra. Both tools have been proven to improve sensitivity of database search results in all types of experiment, particularly endogenous peptides, proteogenomics and metaproteomics.
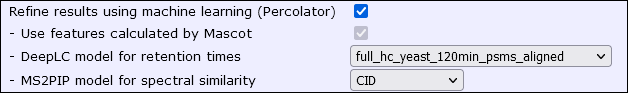
When enabled, the difference between observed and predicted retention time, and the correlation between observed and predicted ion intensities, are combined with core features and used with Percolator rescoring. The results are fully integrated into the Protein Family Summary report and standard export formats. Mascot ships with several pre-trained models for DeepLC and MS2PIP. These are installed locally; Internet access is not required to use MS2Rescore. A GPU is not required.
Nose-to-tail workflow
You can now select refining with machine learning directly in the search form, including DeepLC and MS2PIP models. When enabled, Mascot automatically rescores the results at the end of the database search. We have made a number of improvements to the robustness, speed and logging to make it a smooth experience. When you export results in mzIdentML, mzTab, Mascot CSV or XML formats, the settings are carried over from the search form.
Machine learning quality report
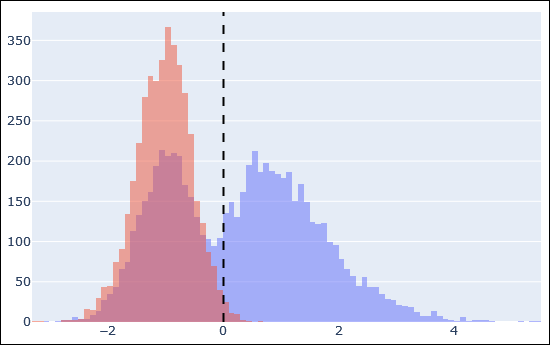
Take the guesswork out of machine learning with the new machine learning quality report. The report helps answering questions like: Did rescoring make the results better? Which features were important? Are the predicted retention times correct? Did I choose the right model for predicted spectra? The report is built on charts and plots provided by MS2Rescore, and it is available with any target-decoy search.
Adapter interface for adding your own ML integration
Mascot Server includes a new adapter interface for accessing peptide feature predictors. The MS2Rescore integration is implemented as an adapter. The interface can be used for any tool that provides feature predictions, and it can be written in any of the programming languages supported by Mascot Parser (C++, C#, Perl, Python, Java). Please contact us if you would like to write an adapter for an in-house tool, or if you would like us to ship an adapter with a future version of Mascot.
Faster and more precise error tolerant search
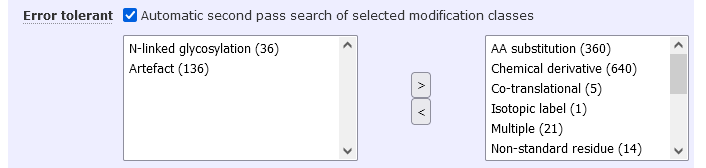
The Mascot Error Tolerant Search is a second pass search that identifies unsuspected chemical and post-translational modifications as well as enzyme non-specificity. You can now restrict the second pass to specific modification classes. For example, search only post-translational modifications, or only N-linked glycosylation.
There are two benefits: Searching only a subset of Unimod drastically reduces the computing time in the second pass of the search; and a smaller search space reduces significance thresholds, which can yield more matches at the same FDR.
Improved results file performance across the system
Mascot versions from 1.0 to 2.8 saved search results in a plain text (MIME format) file. Plain text is great for interoperability, but the size and scale of data sets has now reached a point where the file format is a performance bottleneck. This is especially true for interactive use but also downstream processing, like label-free quantitation in Mascot Distiller.
New file format: Mascot Search Results (MSR)
Mascot Server 3.0 introduces a new file format, Mascot Search Results (MSR). This is a self-contained SQLite database with a formal schema. SQLite is a highly optimised relational database, which provides the foundation for upcoming improvements in Mascot Distiller, as well as improves your experience when browsing and exporting search results.
Backwards compatibility
Every aspect of the system has been re-engineered under the hood, but Mascot Server continues to have strong backwards compatibility:
- View and export all your existing results from any previous version of Mascot Server
- The procedure to submit a search is unaffected by the new file format
- Export MSR file in .dat format if your application or pipeline requires .dat files
- Force Mascot to create a file in .dat format if your application or pipeline reads .dat files directly from the ‘daily’ directory
- The Mascot Server client API is unchanged – continues to support current or older versions of Mascot Daemon, Mascot Distiller, Thermo Proteome Discoverer, etc.
- Mascot Parser has been re-engineered to parse MSR files – over 95% of the API is unchanged
Mascot Daemon improvements
Mascot Server 3.0 ships with the new Mascot Daemon 3.0.
Enable refining with machine learning. The Daemon parameter editor now has controls for enabling refining with machine learning, as well as selecting DeepLC and MS2PIP models.
Search parameters for Unimod classifications with error tolerant searches. The Daemon parameter editor now has controls for selecting modification classifications, same as the Mascot Server search form.
Automatically running reports with the Mascot Daemon Export Extender. Mascot Daemon now ships with a new script, Mascot Daemon Export Extender (MDXE), which automates the steps to generate Distiller quantitation reports.
Report top-3 average intensity proteins in Quantitation Summary. In addition to protein ratios, the report now has the option to calculate the top-3 Average (label-free) intensity for any supported search type.
Allow deleting multiple tasks in one step. When deleting tasks from the Daemon task list, you can now select a range, then delete.
Make peak picking with the Distiller Daemon Toolbox Processor group aware on Windows 11 based systems. When a processor has more than 64 logical cores, Windows splits them into two processor groups. Previously, Daemon was limited to one processor group. On a system with 48 physical cores (96 logical cores), Daemon was using only half the processor (48 logical cores). The new version of Daemon can now use 100% of the processor.
Changes to defaults
Several configuration defaults are changing in this release. We are making these changes to simplify the user experience and reduce the number of choices you need make to get the best results.
Automatic target-decoy search is now the default. The ‘Decoy’ checkbox has been hidden from the search form. Every results report, except spectral library search and intact crosslinking, now displays peptide and protein FDR by default. You can change the default with the AlwaysEnableAutoDecoySearch configuration option.
Sequence FDR is now the default. When you select a target FDR, previous versions of Mascot defaulted to PSM FDR. Mascot Server 3.0 defaults to sequence FDR. You can change the FDR type in Protein Family Summary format controls and when exporting the results. We recommend sequence FDR for all but the smallest data sets.
SplitNumberOfQueries now defaults to 2000. Doubling SplitNumberOfQueries from 1000 to 2000 reduces the duration of most database searches on most systems by 10-15%, at a negligible increase in RAM usage. If your Mascot Server PC has less than 4GB of RAM, you should revert to the old default – but you are better off upgrading your PC and adding more RAM.
Removal of obsolete functionality
Select Summary and Peptide Summary reports are obsolete. These are the reports generated by master_results.pl for MS/MS searches. We encourage everyone to use the Protein Family Summary report, which provides a more accurate picture of your data and supports refining with machine learning. The Select Summary/Peptide Summary reports are still available in Mascot Server 3.0, but they will be removed in a future release.
config/mod_file is obsolete. This file was automatically created from unimod.xml to support legacy client programs. Mascot Server 3.0 no longer creates the file by default. For a transitionary period, the functionality can be re-enabled with a configuration option. The functionality will be permanently removed in Mascot Server 4.0.
Obsolete file formats are no longer selectable in the search form. Support for legacy formats, like Micromass PKL or Sciex API III, is still present but hidden by default. The obsolete search form controls for peptide charge and precursor m/z are now also hidden. The new configuration option SearchSubmitAcceptedFileTypes defaults to MGF and mzML.
The choice for Monoisotopic/Average mass has been removed from the search form. All contemporary data analysis software saves peak lists using monoisotopic masses.
Obsolete export formats are no longer selectable in the export form. Previous versions of Mascot could export results in DTASelect v1.9 and pepXML v1.8 formats. These very old file versions lack support for features added to Mascot in the past decade. You can still select them by using a command-line argument to export_dat_2.pl. DTASelect and pepXML support will be permanently removed in Mascot Server 4.0.
Support for Windows Vista, 7, 8, Server 2008, Server 2008 R2 and Server 2012 has been dropped. On Windows, Mascot Server 3.0 requires Windows 8.1 or later, or Windows Server 2012 R2 or later. We recommend Windows 11/Server 2022.
Support for glibc 2.5 has been dropped. On Linux, Mascot Server 3.0 requires glibc 2.17 or later. You will need a Linux distribution released after 2014. Mascot has no other system dependencies on Linux. We recommend using the latest version of AlmaLinux/CentOS/Rocky Linux, Debian or Ubuntu.
Other improvements
Major update to user documentation. Almost every page in the Mascot HTML help has been reviewed. Dozens of new help pages and tutorials have been added, covering every aspect of peptide and protein identification and quantitation. The full user documentation is shipped with Mascot and freely available on our website.
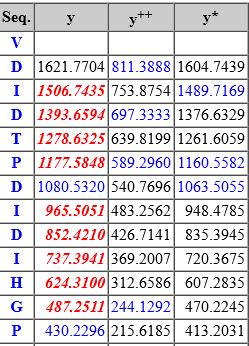
Peptide View now annotates all fragments. The radio button to select between “fragments used for scoring” and “all fragments” has been removed. Fragments used by Mascot for matching and scoring are presented in red, and other fragments (typically at lower peak intensity) are presented in blue.
Reduced “query prep time” on many Windows systems. The time taken to parse large MGF files has been reduced by a factor of 3-4x on most Windows systems. This is the step done at “0%” before the database search has properly started.
Perl has been updated to version 5.38. All Perl modules and support libraries have been updated to the latest versions. This adds TLSv1.3 support to Database Manager. On Windows, Database Manager pages now load much faster thanks to reduced file locking delays.
Javascript speed improvements in the interactive reports. Clicking to expand and collapse content in Protein Family Summary is now smoother. The Javascript code has been switched to the high performing querySelector() API.